Phenotype-information-phenotype cycle for deconvolution of combinatorial antibody libraries selected against complex systems
- PMID: 21825149
- PMCID: PMC3158238
- DOI: 10.1073/pnas.1111218108
Phenotype-information-phenotype cycle for deconvolution of combinatorial antibody libraries selected against complex systems
Abstract
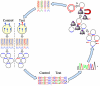
Use of large combinatorial antibody libraries and next-generation sequencing of nucleic acids are two of the most powerful methods in modern molecular biology. The libraries are screened using the principles of evolutionary selection, albeit in real time, to enrich for members with a particular phenotype. This selective process necessarily results in the loss of information about less-fit molecules. On the other hand, sequencing of the library, by itself, gives information that is mostly unrelated to phenotype. If the two methods could be combined, the full potential of very large molecular libraries could be realized. Here we report the implementation of a phenotype-information-phenotype cycle that integrates information and gene recovery. After selection for phage-encoded antibodies that bind to targets expressed on the surface of Escherichia coli, the information content of the selected pool is obtained by pyrosequencing. Sequences that encode specific antibodies are identified by a bioinformatic analysis and recovered by a stringent affinity method that is uniquely suited for gene isolation from a highly degenerate collection of nucleic acids. This approach can be generalized for selection of antibodies against targets that are present as minor components of complex systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Huse WD, et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–1281. - PubMed
-
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

