Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage
- PMID: 21825151
- PMCID: PMC3161609
- DOI: 10.1073/pnas.1108799108
Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage
Abstract
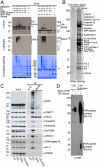
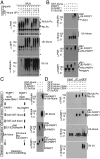
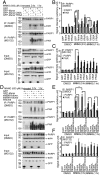
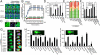
Ubiquitin mediated protein degradation is crucial for regulation of cell signaling and protein quality control. Poly(ADP-ribose) (PAR) is a cell-signaling molecule that mediates changes in protein function through binding at PAR binding sites. Here we characterize the PAR binding protein, Iduna, and show that it is a PAR-dependent ubiquitin E3 ligase. Iduna's E3 ligase activity requires PAR binding because point mutations at Y156A and R157A eliminate Iduna's PAR binding and Iduna's E3 ligase activity. Iduna's E3 ligase activity also requires an intact really interesting new gene (RING) domain because Iduna possessing point mutations at either H54A or C60A is devoid of ubiquitination activity. Tandem affinity purification reveals that Iduna binds to a number of proteins that are either PARsylated or bind PAR including PAR polymerase-1, 2 (PARP1, 2), nucleolin, DNA ligase III, KU70, KU86, XRCC1, and histones. PAR binding to Iduna activates its E3 ligase function, and PAR binding is required for Iduna ubiquitination of PARP1, XRCC1, DNA ligase III, and KU70. Iduna's PAR-dependent ubiquitination of PARP1 targets it for proteasomal degradation. Via PAR binding and ubiquitin E3 ligase activity, Iduna protects against cell death induced by the DNA damaging agent N-methyl-N-nitro-N-nitrosoguanidine (MNNG) and rescues cells from G1 arrest and promotes cell survival after γ-irradiation. Moreover, Iduna facilitates DNA repair by reducing apurinic/apyrimidinic (AP) sites after MNNG exposure and facilitates DNA repair following γ-irradiation as assessed by the comet assay. These results define Iduna as a PAR-dependent E3 ligase that regulates cell survival and DNA repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: A signalling connection. Nat Rev. 2003;4:491–497. - PubMed
-
- Hochstrasser M. New structural clues to substrate specificity in the “ubiquitin system”. Mol Cell. 2002;9:453–454. - PubMed
-
- Harper JW. A phosphorylation-driven ubiquitination switch for cell-cycle control. Trends Cell Biol. 2002;12:104–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

