The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation
- PMID: 21826105
- PMCID: PMC3271587
- DOI: 10.1038/cr.2011.128
The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation
Abstract
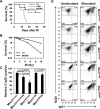
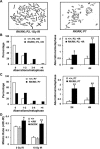
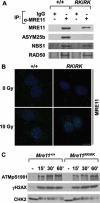
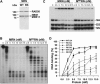
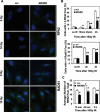
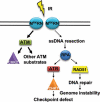
The MRE11/RAD50/NBS1 complex is the primary sensor rapidly recruited to DNA double-strand breaks (DSBs). MRE11 is known to be arginine methylated by PRMT1 within its glycine-arginine-rich (GAR) motif. In this study, we report a mouse knock-in allele of Mre11 that substitutes the arginines with lysines in the GAR motif and generates the MRE11(RK) protein devoid of methylated arginines. The Mre11(RK/RK) mice were hypersensitive to γ-irradiation (IR) and the cells from these mice displayed cell cycle checkpoint defects and chromosome instability. Moreover, the Mre11(RK/RK) MEFs exhibited ATR/CHK1 signaling defects and impairment in the recruitment of RPA and RAD51 to the damaged sites. The M(RK)RN complex formed and localized to the sites of DNA damage and normally activated the ATM pathway in response to IR. The M(RK)RN complex exhibited exonuclease and DNA-binding defects in vitro responsible for the impaired DNA end resection and ATR activation observed in vivo in response to IR. Our findings provide genetic evidence for the critical role of the MRE11 GAR motif in DSB repair, and demonstrate a mechanistic link between post-translational modifications at the MRE11 GAR motif and DSB processing, as well as the ATR/CHK1 checkpoint signaling.
Figures



References
-
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. - PubMed
-
- Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275:19866–19876. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

