Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans--Ignicoccus hospitalis relationship
- PMID: 21826220
- PMCID: PMC3149612
- DOI: 10.1371/journal.pone.0022942
Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans--Ignicoccus hospitalis relationship
Abstract
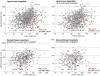
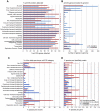
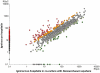
Nanoarchaeum equitans, the only cultured representative of the Nanoarchaeota, is dependent on direct physical contact with its host, the hyperthermophile Ignicoccus hospitalis. The molecular mechanisms that enable this relationship are unknown. Using whole-cell proteomics, differences in the relative abundance of >75% of predicted protein-coding genes from both Archaea were measured to identify the specific response of I. hospitalis to the presence of N. equitans on its surface. A purified N. equitans sample was also analyzed for evidence of interspecies protein transfer. The depth of cellular proteome coverage achieved here is amongst the highest reported for any organism. Based on changes in the proteome under the specific conditions of this study, I. hospitalis reacts to N. equitans by curtailing genetic information processing (replication, transcription) in lieu of intensifying its energetic, protein processing and cellular membrane functions. We found no evidence of significant Ignicoccus biosynthetic enzymes being transported to N. equitans. These results suggest that, under laboratory conditions, N. equitans diverts some of its host's metabolism and cell cycle control to compensate for its own metabolic shortcomings, thus appearing to be entirely dependent on small, transferable metabolites and energetic precursors from I. hospitalis.
Conflict of interest statement
Figures



References
-
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. - PubMed
-
- Jahn U, Summons R, Sturt H, Grosjean E, Huber H. Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I. Arch Microbiol. 2004;182:404–413. - PubMed
-
- Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet. 2009;10:617–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

