Dynamics of the phosphoinositide 3-kinase p110δ interaction with p85α and membranes reveals aspects of regulation distinct from p110α
- PMID: 21827948
- PMCID: PMC3155019
- DOI: 10.1016/j.str.2011.06.003
Dynamics of the phosphoinositide 3-kinase p110δ interaction with p85α and membranes reveals aspects of regulation distinct from p110α
Abstract
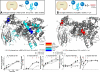
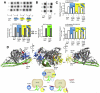
Phosphoinositide 3-kinase δ is upregulated in lymphocytic leukemias. Because the p85-regulatory subunit binds to any class IA subunit, it was assumed there is a single universal p85-mediated regulatory mechanism; however, we find isozyme-specific inhibition by p85α. Using deuterium exchange mass spectrometry (DXMS), we mapped regulatory interactions of p110δ with p85α. Both nSH2 and cSH2 domains of p85α contribute to full inhibition of p110δ, the nSH2 by contacting the helical domain and the cSH2 via the C terminus of p110δ. The cSH2 inhibits p110β and p110δ, but not p110α, implying that p110α is uniquely poised for oncogenic mutations. Binding RTK phosphopeptides disengages the SH2 domains, resulting in exposure of the catalytic subunit. We find that phosphopeptides greatly increase the affinity of the heterodimer for PIP2-containing membranes measured by FRET. DXMS identified regions decreasing exposure at membranes and also regions gaining exposure, indicating loosening of interactions within the heterodimer at membranes.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures





References
-
- Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. - PubMed
-
- Carson J.D., Van Aller G., Lehr R., Sinnamon R.H., Kirkpatrick R.B., Auger K.R., Dhanak D., Copeland R.A., Gontarek R.R., Tummino P.J., Luo L. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem. J. 2008;409:519–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

