Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells
- PMID: 21828039
- PMCID: PMC3190770
- DOI: 10.1074/jbc.M111.280396
Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells
Abstract
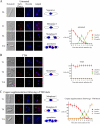
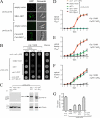
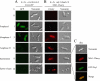
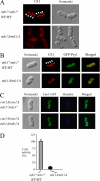
To gain insight in the molecular basis of copper homeostasis during meiosis, we have used DNA microarrays to analyze meiotic gene expression in the model yeast Schizosaccharomyces pombe. Profiling data identified a novel meiosis-specific gene, termed mfc1(+), that encodes a putative major facilitator superfamily-type transporter. Although Mfc1 does not exhibit any significant sequence homology with the copper permease Ctr4, it contains four putative copper-binding motifs that are typically found in members of the copper transporter family of copper transporters. Similarly to the ctr4(+) gene, the transcription of mfc1(+) was induced by low concentrations of copper. However, its temporal expression profile during meiosis was distinct to ctr4(+). Whereas Ctr4 was observed at the plasma membrane shortly after induction of meiosis, Mfc1 appeared later in precursor vesicles and, subsequently, at the forespore membrane of ascospores. Using the fluorescent copper-binding tracker Coppersensor-1 (CS1), labile cellular copper was primarily detected in the forespores in an mfc1(+)/mfc1(+) strain, whereas an mfc1Δ/mfc1Δ mutant exhibited an intracellular dispersed punctate distribution of labile copper ions. In addition, the copper amine oxidase Cao1, which localized primarily in the forespores of asci, was fully active in mfc1(+)/mfc1(+) cells, but its activity was drastically reduced in an mfc1Δ/mfc1Δ strain. Furthermore, our data showed that meiotic cells that express the mfc1(+) gene have a distinct developmental advantage over mfc1Δ/mfc1Δ mutant cells when copper is limiting. Taken together, the data reveal that Mfc1 serves to transport copper for accurate and timely meiotic differentiation under copper-limiting conditions.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

