Shedding of the Mer tyrosine kinase receptor is mediated by ADAM17 protein through a pathway involving reactive oxygen species, protein kinase Cδ, and p38 mitogen-activated protein kinase (MAPK)
- PMID: 21828049
- PMCID: PMC3190938
- DOI: 10.1074/jbc.M111.263020
Shedding of the Mer tyrosine kinase receptor is mediated by ADAM17 protein through a pathway involving reactive oxygen species, protein kinase Cδ, and p38 mitogen-activated protein kinase (MAPK)
Abstract
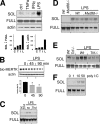
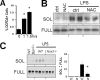
Mer tyrosine kinase (MerTK) is an integral membrane protein that is preferentially expressed by phagocytic cells, where it promotes efferocytosis and inhibits inflammatory signaling. Proteolytic cleavage of MerTK at an unidentified site leads to shedding of its soluble ectodomain (soluble MER; sMER), which can inhibit thrombosis in mice and efferocytosis in vitro. Herein, we show that MerTK is cleaved at proline 485 in murine macrophages. Site-directed deletion of 6 amino acids spanning proline 485 rendered MerTK resistant to proteolysis and suppression of efferocytosis by cleavage-inducing stimuli. LPS is a known inducer of MerTK cleavage, and the intracellular signaling pathways required for this action are unknown. LPS/TLR4-mediated generation of sMER required disintegrin and metalloproteinase ADAM17 and was independent of Myd88, instead requiring TRIF adaptor signaling. LPS-induced cleavage was suppressed by deficiency of NADPH oxidase 2 (Nox2) and PKCδ. The addition of the antioxidant N-acetyl cysteine inhibited PKCδ, and silencing of PKCδ inhibited MAPK p38, which was also required. In a mouse model of endotoxemia, we discovered that LPS induced plasma sMER, and this was suppressed by Adam17 deficiency. Thus, a TRIF-mediated pattern recognition receptor signaling cascade requires NADPH oxidase to activate PKCδ and then p38, culminating in ADAM17-mediated proteolysis of MerTK. These findings link innate pattern recognition receptor signaling to proteolytic inactivation of MerTK and generation of sMER and uncover targets to test how MerTK cleavage affects efferocytosis efficiency and inflammation resolution in vivo.
Figures






Similar articles
-
Angiotensin II deteriorates advanced atherosclerosis by promoting MerTK cleavage and impairing efferocytosis through the AT1R/ROS/p38 MAPK/ADAM17 pathway.Am J Physiol Cell Physiol. 2019 Oct 1;317(4):C776-C787. doi: 10.1152/ajpcell.00145.2019. Epub 2019 Aug 7. Am J Physiol Cell Physiol. 2019. PMID: 31390228
-
Expansion of necrotic core and shedding of Mertk receptor in human carotid plaques: a role for oxidized polyunsaturated fatty acids?Cardiovasc Res. 2013 Jan 1;97(1):125-33. doi: 10.1093/cvr/cvs301. Epub 2012 Sep 20. Cardiovasc Res. 2013. PMID: 22997156
-
Staphylococcus aureus vesicles impair cutaneous wound healing through p38 MAPK-MerTK cleavage-mediated inhibition of macrophage efferocytosis.Cell Commun Signal. 2025 Jan 8;23(1):14. doi: 10.1186/s12964-024-01994-z. Cell Commun Signal. 2025. PMID: 39780180 Free PMC article.
-
TAM receptors and their ligand-mediated activation: Role in atherosclerosis.Int Rev Cell Mol Biol. 2020;357:21-33. doi: 10.1016/bs.ircmb.2020.09.001. Epub 2020 Oct 5. Int Rev Cell Mol Biol. 2020. PMID: 33234243 Free PMC article. Review.
-
Molecular pathways: MERTK signaling in cancer.Clin Cancer Res. 2013 Oct 1;19(19):5275-80. doi: 10.1158/1078-0432.CCR-12-1451. Epub 2013 Jul 5. Clin Cancer Res. 2013. PMID: 23833304 Free PMC article. Review.
Cited by
-
Top Five Stories of the Cellular Landscape and Therapies of Atherosclerosis: Current Knowledge and Future Perspectives.Curr Med Sci. 2024 Feb;44(1):1-27. doi: 10.1007/s11596-023-2818-2. Epub 2023 Dec 7. Curr Med Sci. 2024. PMID: 38057537 Review.
-
2021 Jeffrey M. Hoeg Award Lecture: Defining the Role of Efferocytosis in Cardiovascular Disease: A Focus on the CD47 (Cluster of Differentiation 47) Axis.Arterioscler Thromb Vasc Biol. 2022 Jun;42(6):e145-e154. doi: 10.1161/ATVBAHA.122.317049. Epub 2022 Apr 7. Arterioscler Thromb Vasc Biol. 2022. PMID: 35387480 Free PMC article. Review.
-
Crosstalk Between Macrophages and Vascular Smooth Muscle Cells in Atherosclerotic Plaque Stability.Arterioscler Thromb Vasc Biol. 2022 Apr;42(4):372-380. doi: 10.1161/ATVBAHA.121.316233. Epub 2022 Feb 17. Arterioscler Thromb Vasc Biol. 2022. PMID: 35172605 Free PMC article. Review.
-
The role of non-resolving inflammation in atherosclerosis.J Clin Invest. 2018 Jul 2;128(7):2713-2723. doi: 10.1172/JCI97950. Epub 2018 Jul 2. J Clin Invest. 2018. PMID: 30108191 Free PMC article. Review.
-
The regulation and function of acetylated high-mobility group box 1 during implantation and decidualization.Front Immunol. 2023 Jan 25;14:1024706. doi: 10.3389/fimmu.2023.1024706. eCollection 2023. Front Immunol. 2023. PMID: 36761729 Free PMC article.
References
-
- Nagata K., Ohashi K., Nakano T., Arita H., Zong C., Hanafusa H., Mizuno K. (1996) J. Biol. Chem. 271, 30022–30027 - PubMed
-
- Uehara H., Shacter E. (2008) J. Immunol. 180, 2522–2530 - PubMed
-
- Scott R. S., McMahon E. J., Pop S. M., Reap E. A., Caricchio R., Cohen P. L., Earp H. S., Matsushima G. K. (2001) Nature 411, 207–211 - PubMed
-
- Chen C., Li Q., Darrow A. L., Wang Y., Derian C. K., Yang J., de Garavilla L., Andrade-Gordon P., Damiano B. P. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1118–1123 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

