Characterization of the dimerization interface of membrane type 4 (MT4)-matrix metalloproteinase
- PMID: 21828052
- PMCID: PMC3190889
- DOI: 10.1074/jbc.M111.253369
Characterization of the dimerization interface of membrane type 4 (MT4)-matrix metalloproteinase
Abstract
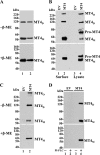
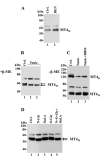
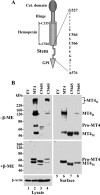
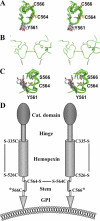
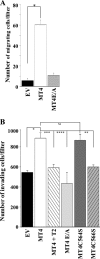
MT4-MMP (MMP17) belongs to a unique subset of membrane type-matrix metalloproteinases that are anchored to the cell surface via a glycosylphosphatidylinositol moiety. However, little is known about its biochemical properties. Here, we report that MT4-MMP is displayed on the cell surface as a mixed population of monomeric, dimeric, and oligomeric forms. Sucrose gradient fractionation demonstrated that these forms of MT4-MMP are all present in lipid rafts. Mutational and computational analyses revealed that Cys(564), which is present within the stem region, mediates MT4-MMP homodimerization by forming a disulfide bond. Substitution of Cys(564) results in a more rapid MT4-MMP turnover, when compared with the wild-type enzyme, consistent with a role for dimerization in protein stability. Expression of MT4-MMP in Madin-Darby canine kidney cells enhanced cell migration and invasion of Matrigel, a process that requires catalytic activity. However, a serine substitution at Cys(564) did not reduce MT4-MMP-stimulated cell invasion of Matrigel suggesting that homodimerization is not required for this process. Deglycosylation studies showed that MT4-MMP is modified by N-glycosylation. Moreover, inhibition of N-glycosylation by tunicamycin diminished the extent of MT4-MMP dimerization suggesting that N-glycans may confer stability to the dimeric form. Taken together, the data presented here provide a new insight into the characteristics of MT4-MMP and highlight the common and distinct properties of the glycosylphosphatidylinositol-anchored membrane type-matrix metalloproteinases.
Figures


Similar articles
-
Identification and role of the homodimerization interface of the glycosylphosphatidylinositol-anchored membrane type 6 matrix metalloproteinase (MMP25).J Biol Chem. 2008 Dec 12;283(50):35023-32. doi: 10.1074/jbc.M806553200. Epub 2008 Oct 20. J Biol Chem. 2008. PMID: 18936094 Free PMC article.
-
Expression and Characterization of Membrane-Type 4 Matrix Metalloproteinase (MT4-MMP) and its Different Forms in Melanoma.Cell Physiol Biochem. 2017;42(1):198-210. doi: 10.1159/000477311. Epub 2017 May 25. Cell Physiol Biochem. 2017. PMID: 28531887
-
Dynamics of internalization and recycling of the prometastatic membrane type 4 matrix metalloproteinase (MT4-MMP) in breast cancer cells.FEBS J. 2016 Feb;283(4):704-22. doi: 10.1111/febs.13625. Epub 2016 Jan 4. FEBS J. 2016. PMID: 26663028
-
Molecular Mechanisms Driven by MT4-MMP in Cancer Progression.Int J Mol Sci. 2023 Jun 9;24(12):9944. doi: 10.3390/ijms24129944. Int J Mol Sci. 2023. PMID: 37373092 Free PMC article. Review.
-
MT4-MMP: The GPI-Anchored Membrane-Type Matrix Metalloprotease with Multiple Functions in Diseases.Int J Mol Sci. 2019 Jan 16;20(2):354. doi: 10.3390/ijms20020354. Int J Mol Sci. 2019. PMID: 30654475 Free PMC article. Review.
Cited by
-
Placental membrane-type metalloproteinases (MT-MMPs): Key players in pregnancy.Cell Adh Migr. 2016 Mar 3;10(1-2):136-46. doi: 10.1080/19336918.2015.1110671. Epub 2016 Jan 8. Cell Adh Migr. 2016. PMID: 26745344 Free PMC article. Review.
-
Post-Translational Modification-Dependent Activity of Matrix Metalloproteinases.Int J Mol Sci. 2019 Jun 24;20(12):3077. doi: 10.3390/ijms20123077. Int J Mol Sci. 2019. PMID: 31238509 Free PMC article. Review.
-
Glycosylation of matrix metalloproteases and tissue inhibitors: present state, challenges and opportunities.Biochem J. 2016 Jun 1;473(11):1471-82. doi: 10.1042/BJ20151154. Biochem J. 2016. PMID: 27234584 Free PMC article. Review.
-
Drug Development for Metastasis Prevention.Crit Rev Oncog. 2015;20(5-6):449-73. doi: 10.1615/CritRevOncog.v20.i5-6.150. Crit Rev Oncog. 2015. PMID: 27279241 Free PMC article. Review.
-
Identification of a high-mannose ICAM-1 glycoform: effects of ICAM-1 hypoglycosylation on monocyte adhesion and outside in signaling.Am J Physiol Cell Physiol. 2013 Jul 15;305(2):C228-37. doi: 10.1152/ajpcell.00116.2013. Epub 2013 May 22. Am J Physiol Cell Physiol. 2013. PMID: 23703526 Free PMC article.
References
-
- Zucker S., Pei D., Cao J., Lopez-Otin C. (2003) Curr. Top. Dev. Biol. 54, 1–74 - PubMed
-
- English W. R., Puente X. S., Freije J. M., Knauper V., Amour A., Merryweather A., Lopez-Otin C., Murphy G. (2000) J. Biol. Chem. 275, 14046–14055 - PubMed
-
- English W. R., Velasco G., Stracke J. O., Knäuper V., Murphy G. (2001) FEBS Lett. 491, 137–142 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

