Cell of origin strongly influences genetic selection in a mouse model of T-ALL
- PMID: 21828136
- PMCID: PMC3208280
- DOI: 10.1182/blood-2011-03-343947
Cell of origin strongly influences genetic selection in a mouse model of T-ALL
Abstract
Identifying the normal cell from which a tumor originates is crucial to understanding the etiology of that cancer. However, retrospective identification of the cell of origin in cancer is challenging because of the accumulation of genetic and epigenetic changes in tumor cells. The biologic state of the cell of origin likely influences the genetic events that drive transformation. We directly tested this hypothesis by performing a Sleeping Beauty transposon mutagenesis screen in which common insertion sites were identified in tumors that were produced by mutagenesis of cells at varying time points throughout the T lineage. Mutation and gene expression data derived from these tumors were then compared with data obtained from a panel of 84 human T-cell acute lymphoblastic leukemia samples, including copy number alterations and gene expression profiles. This revealed that altering the cell of origin produces tumors that model distinct subtypes of human T-cell acute lymphoblastic leukemia, suggesting that even subtle changes in the cell of origin dramatically affect genetic selection in tumors. These findings have broad implications for the genetic analysis of human cancers as well as the production of mouse models of cancer.
Figures

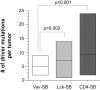
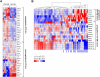
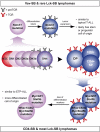
Comment in
-
Sleeping beauty: does ETP-ALL awaken later?Blood. 2011 Oct 27;118(17):4500-1. doi: 10.1182/blood-2011-09-375121. Blood. 2011. PMID: 22033941 No abstract available.
References
-
- Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. - PubMed
-
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436(7048):221–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

