DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation
- PMID: 21828137
- PMCID: PMC3186332
- DOI: 10.1182/blood-2011-06-357996
DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation
Abstract
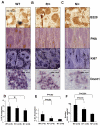
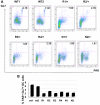
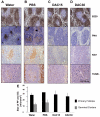
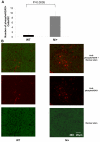
The phenotype of germinal center (GC) B cells includes the unique ability to tolerate rapid proliferation and the mutagenic actions of activation induced cytosine deaminase (AICDA). Given the importance of epigenetic patterning in determining cellular phenotypes, we examined DNA methylation and the role of DNA methyltransferases in the formation of GCs. DNA methylation profiling revealed a marked shift in DNA methylation patterning in GC B cells versus resting/naive B cells. This shift included significant differential methylation of 235 genes, with concordant inverse changes in gene expression affecting most notably genes of the NFkB and MAP kinase signaling pathways. GC B cells were predominantly hypomethylated compared with naive B cells and AICDA binding sites were highly overrepresented among hypomethylated loci. GC B cells also exhibited greater DNA methylation heterogeneity than naive B cells. Among DNA methyltransferases (DNMTs), only DNMT1 was significantly up-regulated in GC B cells. Dnmt1 hypomorphic mice displayed deficient GC formation and treatment of mice with the DNA methyltransferase inhibitor decitabine resulted in failure to form GCs after immune stimulation. Notably, the GC B cells of Dnmt1 hypomorphic animals showed evidence of increased DNA damage, suggesting dual roles for DNMT1 in DNA methylation and double strand DNA break repair.
Figures



References
-
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. - PubMed
-
- de Yebenes VG, Ramiro AR. Activation-induced deaminase: light and dark sides. Trends Mol Med. 2006;12(9):432–439. - PubMed
-
- Martin A, Bardwell PD, Woo CJ, Fan M, Shulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;415(6873):802–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

