Genome engineering with zinc-finger nucleases
- PMID: 21828278
- PMCID: PMC3176093
- DOI: 10.1534/genetics.111.131433
Genome engineering with zinc-finger nucleases
Abstract
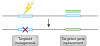
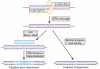
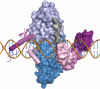
Zinc-finger nucleases (ZFNs) are targetable DNA cleavage reagents that have been adopted as gene-targeting tools. ZFN-induced double-strand breaks are subject to cellular DNA repair processes that lead to both targeted mutagenesis and targeted gene replacement at remarkably high frequencies. This article briefly reviews the history of ZFN development and summarizes applications that have been made to genome editing in many different organisms and situations. Considerable progress has been made in methods for deriving zinc-finger sets for new genomic targets, but approaches to design and selection are still being perfected. An issue that needs more attention is the extent to which available mechanisms of double-strand break repair limit the scope and utility of ZFN-initiated events. The bright prospects for future applications of ZFNs, including human gene therapy, are discussed.
Figures

References
-
- Alwin S., Gere M. B., Gulh E., Effertz K., Barbas C. F., III, et al. , 2005. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 12: 610–617 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

