Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication
- PMID: 21828280
- PMCID: PMC3176089
- DOI: 10.1534/genetics.111.127324
Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication
Abstract
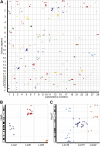
Genomic resources for hundreds of species of evolutionary, agricultural, economic, and medical importance are unavailable due to the expense of well-assembled genome sequences and difficulties with multigenerational studies. Teleost fish provide many models for human disease but possess anciently duplicated genomes that sometimes obfuscate connectivity. Genomic information representing a fish lineage that diverged before the teleost genome duplication (TGD) would provide an outgroup for exploring the mechanisms of evolution after whole-genome duplication. We exploited massively parallel DNA sequencing to develop meiotic maps with thrift and speed by genotyping F(1) offspring of a single female and a single male spotted gar (Lepisosteus oculatus) collected directly from nature utilizing only polymorphisms existing in these two wild individuals. Using Stacks, software that automates the calling of genotypes from polymorphisms assayed by Illumina sequencing, we constructed a map containing 8406 markers. RNA-seq on two map-cross larvae provided a reference transcriptome that identified nearly 1000 mapped protein-coding markers and allowed genome-wide analysis of conserved synteny. Results showed that the gar lineage diverged from teleosts before the TGD and its genome is organized more similarly to that of humans than teleosts. Thus, spotted gar provides a critical link between medical models in teleost fish, to which gar is biologically similar, and humans, to which gar is genomically similar. Application of our F(1) dense mapping strategy to species with no prior genome information promises to facilitate comparative genomics and provide a scaffold for ordering the numerous contigs arising from next generation genome sequencing.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

