The structure of a transcription activation subcomplex reveals how σ(70) is recruited to PhoB promoters
- PMID: 21829166
- PMCID: PMC3173795
- DOI: 10.1038/emboj.2011.271
The structure of a transcription activation subcomplex reveals how σ(70) is recruited to PhoB promoters
Abstract
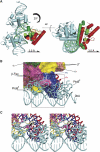
PhoB is a two-component response regulator that activates transcription by interacting with the σ(70) subunit of the E. coli RNA polymerase in promoters in which the -35 σ(70)-recognition element is replaced by the pho box. The crystal structure of a transcription initiation subcomplex that includes the σ(4) domain of σ(70) fused with the RNA polymerase β subunit flap tip helix, the PhoB effector domain and the pho box DNA reveals how σ(4) recognizes the upstream pho box repeat. As with the -35 element, σ(4) achieves this recognition through the N-terminal portion of its DNA recognition helix, but contact with the DNA major groove is less extensive. Unexpectedly, the same recognition helix contacts the transactivation loop and helices α2 and α3 of PhoB. This result shows a simple and elegant mechanism for polymerase recruitment to pho box promoters in which the lost -35 element contacts are compensated by new ones with the activator. In addition, σ(4) is reoriented, thereby suggesting a remodelling mechanism for transcription initiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

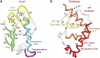



References
-
- Abrahams JP, Leslie AG (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr 52: 30–42 - PubMed
-
- Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH (2002) Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297: 1562–1566 - PubMed
-
- Blanco AG, Solà M, Gomis-Rüth FX, Coll M (2002) Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10: 701–713 - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 - PubMed
-
- Burgess RR (1969) Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem 244: 6168–6176 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

