Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression
- PMID: 21829383
- PMCID: PMC3145624
- DOI: 10.1371/journal.pgen.1002206
Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression
Abstract
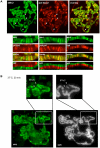
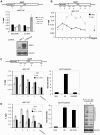
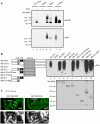
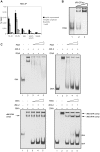
Eukaryotic cells respond to genomic and environmental stresses, such as DNA damage and heat shock (HS), with the synthesis of poly-[ADP-ribose] (PAR) at specific chromatin regions, such as DNA breaks or HS genes, by PAR polymerases (PARP). Little is known about the role of this modification during cellular stress responses. We show here that the nucleosome remodeler dMi-2 is recruited to active HS genes in a PARP-dependent manner. dMi-2 binds PAR suggesting that this physical interaction is important for recruitment. Indeed, a dMi-2 mutant unable to bind PAR does not localise to active HS loci in vivo. We have identified several dMi-2 regions which bind PAR independently in vitro, including the chromodomains and regions near the N-terminus containing motifs rich in K and R residues. Moreover, upon HS gene activation, dMi-2 associates with nascent HS gene transcripts, and its catalytic activity is required for efficient transcription and co-transcriptional RNA processing. RNA and PAR compete for dMi-2 binding in vitro, suggesting a two step process for dMi-2 association with active HS genes: initial recruitment to the locus via PAR interaction, followed by binding to nascent RNA transcripts. We suggest that stress-induced chromatin PARylation serves to rapidly attract factors that are required for an efficient and timely transcriptional response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

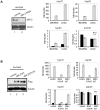
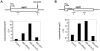
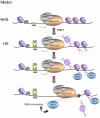
References
-
- Winegarden NA, Wong KS, Sopta M, Westwood JT. Sodium salicylate decreases intracellular ATP, induces both heat shock factor binding and chromosomal puffing, but does not induce hsp 70 gene transcription in Drosophila. J Biol Chem. 1996;271:26971–26980. - PubMed
-
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

