A strategy for full interrogation of prognostic gene expression patterns: exploring the biology of diffuse large B cell lymphoma
- PMID: 21829609
- PMCID: PMC3150354
- DOI: 10.1371/journal.pone.0022267
A strategy for full interrogation of prognostic gene expression patterns: exploring the biology of diffuse large B cell lymphoma
Abstract
Background: Gene expression profiling yields quantitative data on gene expression used to create prognostic models that accurately predict patient outcome in diffuse large B cell lymphoma (DLBCL). Often, data are analyzed with genes classified by whether they fall above or below the median expression level. We sought to determine whether examining multiple cut-points might be a more powerful technique to investigate the association of gene expression with outcome.
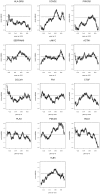
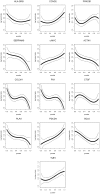
Methodology/principal findings: We explored gene expression profiling data using variable cut-point analysis for 36 genes with reported prognostic value in DLBCL. We plotted two-group survival logrank test statistics against corresponding cut-points of the gene expression levels and smooth estimates of the hazard ratio of death versus gene expression levels. To facilitate comparisons we also standardized the expression of each of the genes by the fraction of patients that would be identified by any cut-point. A multiple comparison adjusted permutation p-value identified 3 different patterns of significance: 1) genes with significant cut-point points below the median, whose loss is associated with poor outcome (e.g. HLA-DR); 2) genes with significant cut-points above the median, whose over-expression is associated with poor outcome (e.g. CCND2); and 3) genes with significant cut-points on either side of the median, (e.g. extracellular molecules such as FN1).
Conclusions/significance: Variable cut-point analysis with permutation p-value calculation can be used to identify significant genes that would not otherwise be identified with median cut-points and may suggest biological patterns of gene effects.
Conflict of interest statement
Figures
References
-
- Roberts RA, Sabalos CM, LeBlanc ML, Martel RR, Frutiger YM, et al. Quantitative nuclease protection assay in paraffin-embedded tissue replicates prognostic microarray gene expression in diffuse large-B-cell lymphoma. Laboratory Investigation. 2007;87:979–997. - PubMed
-
- Chang KC, Huang GC, Jones D, Lin YH. Distribution patterns of dendritic cells and T cells in diffuse large B-cell lymphomas correlate with prognoses. Clin Cancer Res. 2007;13:6666–6672. - PubMed
-
- Dave SS, Fu K, Wright GW, Lam LT, Kluin P, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. - PubMed
-
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. New England Journal of Medicine. 2006;354:2419–2430. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

