Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch's membrane via cleavage of extracellular matrix components
- PMID: 21829675
- PMCID: PMC3149070
- DOI: 10.1371/journal.pone.0022959
Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch's membrane via cleavage of extracellular matrix components
Abstract
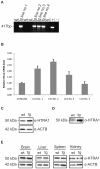
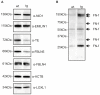
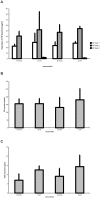
Variants in the chromosomal region 10q26 are strongly associated with an increased risk for age-related macular degeneration (AMD). Two potential AMD genes are located in this region: ARMS2 and HTRA1 (high-temperature requirement A1). Previous studies have suggested that polymorphisms in the promotor region of HTRA1 result in overexpression of HTRA1 protein. This study investigated the role of HTRA1 overexpression in the pathogenesis of AMD. Transgenic Htra1 mice overexpressing the murine protein in the retinal pigment epithelium (RPE) layer of the retina were generated and characterized by transmission electron microscopy, immunofluorescence staining and Western Blot analysis. The elastic layer of Bruch's membrane (BM) in the Htra1 transgenic mice was fragmented and less continuous than in wild type (WT) controls. Recombinant HTRA1 lacking the N-terminal domain cleaved various extracellular matrix (ECM) proteins. Subsequent Western Blot analysis revealed an overexpression of fibronectin fragments and a reduction of fibulin 5 and tropoelastin in the RPE/choroid layer in transgenic mice compared to WT. Fibulin 5 is essential for elastogenesis by promoting elastic fiber assembly and maturation. Taken together, our data implicate that HTRA1 overexpression leads to an altered elastogenesis in BM through fibulin 5 cleavage. It highlights the importance of ECM related proteins in the development of AMD and links HTRA1 to other AMD risk genes such as fibulin 5, fibulin 6, ARMS2 and TIMP3.
Conflict of interest statement
Figures





References
-
- Thornton J, Edwards R, Mitchell P, Harrison RA, Buchan I, et al. Smoking and age-related macular degeneration: a review of association. Eye (Lond) 2005;19:935–944. - PubMed
-
- Fisher SA, Abecasis GR, Yashar BM, Zareparsi S, Swaroop A, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–2264. - PubMed
-
- Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

