Overexpression of a cytosolic pyrophosphatase (TgPPase) reveals a regulatory role of PP(i) in glycolysis for Toxoplasma gondii
- PMID: 21831041
- PMCID: PMC4874478
- DOI: 10.1042/BJ20110641
Overexpression of a cytosolic pyrophosphatase (TgPPase) reveals a regulatory role of PP(i) in glycolysis for Toxoplasma gondii
Abstract
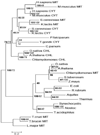
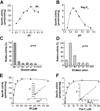
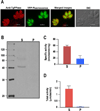
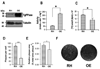
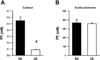
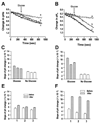
PP(i) is a critical element of cellular metabolism as both an energy donor and as an allosteric regulator of several metabolic pathways. The apicomplexan parasite Toxoplasma gondii uses PP(i) in place of ATP as an energy donor in at least two reactions: the glycolytic PP(i)-dependent PFK (phosphofructokinase) and V-H(+)-PPase [vacuolar H(+)-translocating PPase (pyrophosphatase)]. In the present study, we report the cloning, expression and characterization of cytosolic TgPPase (T. gondii soluble PPase). Amino acid sequence alignment and phylogenetic analysis indicates that the gene encodes a family I soluble PPase. Overexpression of the enzyme in extracellular tachyzoites led to a 6-fold decrease in the cytosolic concentration of PP(i) relative to wild-type strain RH tachyzoites. Unexpectedly, this subsequent reduction in PP(i) was associated with a higher glycolytic flux in the overexpressing mutants, as evidenced by higher rates of proton and lactate extrusion. In addition to elevated glycolytic flux, TgPPase-overexpressing tachyzoites also possessed higher ATP concentrations relative to wild-type RH parasites. These results implicate PP(i) as having a significant regulatory role in glycolysis and, potentially, other downstream processes that regulate growth and cell division.
Figures


References
-
- Kornberg A. On the metabolic significance of phosphorolytic and pyrophosphorolytic reactions. In: Kasha HaPB., editor. Horizons in Biochemistry. New York: Academic Press, Inc.; 1962. pp. 251–264.
-
- Mansurova SE. Inorganic pyrophosphate in mitochondrial metabolism. Biochim Biophys Acta. 1989;977:237–247. - PubMed
-
- Veech RL, Cook GA, King MT. Relationship of free cytoplasmic pyrophosphate to liver glucose content and total pyrophosphate to cytoplasmic phosphorylation potential. FEBS Lett. 1980;117(Suppl):K65–K72. - PubMed
-
- Baltscheffsky M, Schultz A, Baltscheffsky H. H+ -PPases: a tightly membrane-bound family. FEBS Lett. 1999;457:527–533. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

