Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression
- PMID: 21831286
- PMCID: PMC3236340
- DOI: 10.1186/bcr2924
Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression
Abstract
Introduction: Altered expression of Septin 9 (SEPT9), a septin coding for multiple isoform variants, has been observed in several carcinomas, including colorectal, head and neck, ovarian and breast, compared to normal tissues. The mechanisms regulating its expression during tumor initiation and progression in vivo and the oncogenic function of its different isoforms remain elusive.
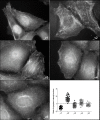
Methods: Using an integrative approach, we investigated SEPT9 at the genetic, epigenetic, mRNA and protein levels in breast cancer. We analyzed a panel of breast cancer cell lines, human primary tumors and corresponding tumor-free areas, normal breast tissues from reduction mammoplasty patients, as well as primary mammary gland adenocarcinomas derived from the polyoma virus middle T antigen, or PyMT, mouse model. MCF7 clones expressing individual GFP-tagged SEPT9 isoforms were used to determine their respective intracellular distributions and effects on cell migration.
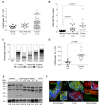
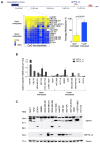
Results: An overall increase in gene amplification and altered expression of SEPT9 were observed during breast tumorigenesis. We identified an intragenic alternative promoter at which methylation regulates SEPT9_v3 expression. Transfection of specific GFP-SEPT9 isoforms in MCF7 cells indicates that these isoforms exhibit differential localization and affect migration rates. Additionally, the loss of an uncharacterized SEPT9 nucleolar localization is observed during tumorigenesis.
Conclusions: In this study, we found conserved in vivo changes of SEPT9 gene amplification and overexpression during human and mouse breast tumorigenesis. We show that DNA methylation is a prominent mechanism responsible for regulating differential SEPT9 isoform expression and that breast tumor samples exhibit distinctive SEPT9 intracellular localization. Together, these findings support the significance of SEPT9 as a promising tool in breast cancer detection and further emphasize the importance of analyzing and targeting SEPT9 isoform-specific expression and function.
Figures


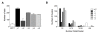
Comment in
-
Steps solidifying a role for SEPT9 in breast cancer suggest that greater strides are needed.Breast Cancer Res. 2012 Jan 9;14(1):101. doi: 10.1186/bcr3056. Breast Cancer Res. 2012. PMID: 22236777 Free PMC article.
References
-
- Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z, Ried T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63:2179–2187. - PubMed
-
- Sørensen AB, Lund AH, Ethelberg S, Copeland NG, Jenkins NA, Pedersen FS. Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family. J Virol. 2000;74:2161–2168. doi: 10.1128/JVI.74.5.2161-2168.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

