Catalysis by dihydrofolate reductase and other enzymes arises from electrostatic preorganization, not conformational motions
- PMID: 21831831
- PMCID: PMC3161549
- DOI: 10.1073/pnas.1111252108
Catalysis by dihydrofolate reductase and other enzymes arises from electrostatic preorganization, not conformational motions
Abstract
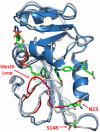
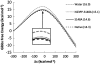
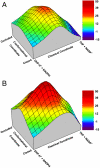
The proposal that enzymatic catalysis is due to conformational fluctuations has been previously promoted by means of indirect considerations. However, recent works have focused on cases where the relevant motions have components toward distinct conformational regions, whose population could be manipulated by mutations. In particular, a recent work has claimed to provide direct experimental evidence for a dynamical contribution to catalysis in dihydrofolate reductase, where blocking a relevant conformational coordinate was related to the suppression of the motion toward the occluded conformation. The present work utilizes computer simulations to elucidate the true molecular basis for the experimentally observed effect. We start by reproducing the trend in the measured change in catalysis upon mutations (which was assumed to arise as a result of a "dynamical knockout" caused by the mutations). This analysis is performed by calculating the change in the corresponding activation barriers without the need to invoke dynamical effects. We then generate the catalytic landscape of the enzyme and demonstrate that motions in the conformational space do not help drive catalysis. We also discuss the role of flexibility and conformational dynamics in catalysis, once again demonstrating that their role is negligible and that the largest contribution to catalysis arises from electrostatic preorganization. Finally, we point out that the changes in the reaction potential surface modify the reorganization free energy (which includes entropic effects), and such changes in the surface also alter the corresponding motion. However, this motion is never the reason for catalysis, but rather simply a reflection of the shape of the reaction potential surface.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
"Eppur si muove" (Yet it moves).Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):15013-4. doi: 10.1073/pnas.1112014108. Epub 2011 Sep 1. Proc Natl Acad Sci U S A. 2011. PMID: 21885737 Free PMC article. No abstract available.
References
-
- Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. - PubMed
-
- Epstein DM, Benkovic SJ, Wright PE. Dynamics of the dihydrofolate-reductase folate complex—catalytic sites and regions known to undergo conformational change exhibit diverse dynamical features. Biochemistry. 1995;34:11037–11048. - PubMed
-
- Cameron CE, Benkovic SJ. Evidence for a functional role of the dynamics of glycine-121 of Escherichia coli dihydrofolate reductase obtained from kinetic analysis of a site-directed mutant. Biochemistry. 1997;36:15792–15800. - PubMed
-
- Henzler-Wildman KA, et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

