Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR
- PMID: 21832022
- PMCID: PMC3187289
- DOI: 10.1128/JCM.00488-11
Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR
Abstract
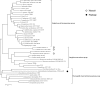
Although staphylococci are identified by phenotypic analysis in many clinical laboratories, these results are often incorrect because of phenotypic variation. Genetic analysis is necessary for definitive species identification. In the present study, we developed a simple multiplex-PCR (M-PCR) for species identification of human-associated staphylococci, which were as follows: Staphylococcus aureus, S. capitis, S. caprae, S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, S. saprophyticus, and S. warneri. This method was designed on the basis of nucleotide sequences of the thermonuclease (nuc) genes that were universally conserved in staphylococci except the S. sciuri group and showed moderate sequence diversity. In order to validate this assay, 361 staphylococcal strains were studied, which had been identified at the species levels by sequence analysis of the hsp60 genes. In consequence, M-PCR demonstrated a sensitivity of 100% and a specificity of 100%. By virtue of simplicity and accuracy, this method will be useful in clinical research.
Figures

References
-
- Bizzini A., Durussel C., Bille J., Greub G., Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 - PMC - PubMed
-
- Carretto E., et al. 2005. Identification of coagulase-negative staphylococci other than Staphylococcus epidermidis by automated ribotyping. Clin. Microbiol. Infect. 11:177–184 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

