Truncation of murine CaV1.2 at Asp-1904 results in heart failure after birth
- PMID: 21832054
- PMCID: PMC3190806
- DOI: 10.1074/jbc.M111.252312
Truncation of murine CaV1.2 at Asp-1904 results in heart failure after birth
Abstract
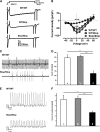
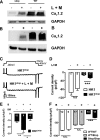
The carboxyl-terminal intracellular tail of the L-type Ca(2+) channel CaV1.2 modulates various aspects of channel activity.For example, deletion of the carboxyl-terminal sequence at Ser-1905 increased CaV1.2 currents in an expression model. To verify this finding in an animal model, we inserted three stop codons at the corresponding Asp-1904 in the murine CaV1.2 gene. Mice homozygous for the Stop mutation (Stop/Stop mice)were born at a Mendelian ratio but died after birth. Stop/Stop hearts showed reduced beating frequencies and contractions.Surprisingly, Stop/Stop cardiomyocytes displayed reduced IBa and a minor expression of the CaV1.2Stop protein. In contrast,expression of the CaV1.2Stop protein was normal in pooled smooth muscle samples from Stop/Stop embryos. As the CaV1.2 channel exists in a cardiac and smooth muscle splice variant, HK1 and LK1, respectively, we analyzed the consequences of the deletion of the carboxyl terminus in the respective splice variant using the rabbit CaV1.2 clone expressed in HEK293 cells.HEK293 cells transfected with the HK1Stop channel showed a reduced IBa and CaV1.2 expression. Treatment with proteasome inhibitors increased the expression of HK1Stop protein and IBa in HEK293 cells and in Stop/Stop cardiomyocytes indicating that truncation of CaV1.2 containing the cardiac exon 1a amino terminus results in proteasomal degradation of the translated protein. In contrast, HEK293 cells transfected with the LK1Stop channel had normal IBa and CaV1.2 expression. These findings indicate that absence of the carboxyl-terminal tail differentially determines the fate of the cardiac and smooth muscle splice variant of the CaV1.2 channel in the mouse.
Figures




Similar articles
-
Truncation of murine CaV1.2 at Asp 1904 increases CaV1.3 expression in embryonic atrial cardiomyocytes.Pflugers Arch. 2013 Jul;465(7):955-64. doi: 10.1007/s00424-012-1212-x. Epub 2013 Jan 22. Pflugers Arch. 2013. PMID: 23338940
-
Exclusion of alternative exon 33 of CaV1.2 calcium channels in heart is proarrhythmogenic.Proc Natl Acad Sci U S A. 2017 May 23;114(21):E4288-E4295. doi: 10.1073/pnas.1617205114. Epub 2017 May 10. Proc Natl Acad Sci U S A. 2017. PMID: 28490495 Free PMC article.
-
Regulation of Blood Pressure by Targeting CaV1.2-Galectin-1 Protein Interaction.Circulation. 2018 Oct 2;138(14):1431-1445. doi: 10.1161/CIRCULATIONAHA.117.031231. Circulation. 2018. PMID: 29650545 Free PMC article.
-
Tissue-specific adrenergic regulation of the L-type Ca2+ channel CaV1.2.Sci Signal. 2020 Dec 22;13(663):eabc6438. doi: 10.1126/scisignal.abc6438. Sci Signal. 2020. PMID: 33443233 Free PMC article. Review.
-
L-type CaV1.2 calcium channels: from in vitro findings to in vivo function.Physiol Rev. 2014 Jan;94(1):303-26. doi: 10.1152/physrev.00016.2013. Physiol Rev. 2014. PMID: 24382889 Review.
Cited by
-
Trafficking highways to the intercalated disc: new insights unlocking the specificity of connexin 43 localization.Cell Commun Adhes. 2014 Feb;21(1):43-54. doi: 10.3109/15419061.2013.876014. Cell Commun Adhes. 2014. PMID: 24460200 Free PMC article. Review.
-
Protein kinase C enhances plasma membrane expression of cardiac L-type calcium channel, CaV1.2.Channels (Austin). 2017 Nov 2;11(6):604-615. doi: 10.1080/19336950.2017.1369636. Epub 2017 Sep 21. Channels (Austin). 2017. PMID: 28901828 Free PMC article.
-
Basal and β-adrenergic regulation of the cardiac calcium channel CaV1.2 requires phosphorylation of serine 1700.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16598-603. doi: 10.1073/pnas.1419129111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368181 Free PMC article.
-
Truncation of murine CaV1.2 at Asp 1904 increases CaV1.3 expression in embryonic atrial cardiomyocytes.Pflugers Arch. 2013 Jul;465(7):955-64. doi: 10.1007/s00424-012-1212-x. Epub 2013 Jan 22. Pflugers Arch. 2013. PMID: 23338940
-
Manipulating L-type calcium channels in cardiomyocytes using split-intein protein transsplicing.Proc Natl Acad Sci U S A. 2013 Sep 17;110(38):15461-6. doi: 10.1073/pnas.1308161110. Epub 2013 Sep 3. Proc Natl Acad Sci U S A. 2013. PMID: 24003157 Free PMC article.
References
-
- Catterall W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521–555 - PubMed
-
- Seisenberger C., Specht V., Welling A., Platzer J., Pfeifer A., Kühbandner S., Striessnig J., Klugbauer N., Feil R., Hofmann F. (2000) J. Biol. Chem. 275, 39193–39199 - PubMed
-
- Wegener J. W., Schulla V., Lee T. S., Koller A., Feil S., Feil R., Kleppisch T., Klugbauer N., Moosmang S., Welling A., Hofmann F. (2004) FASEB J. 18, 1159–1161 - PubMed
-
- Wegener J. W., Schulla V., Koller A., Klugbauer N., Feil R., Hofmann F. (2006) FASEB J. 20, 1260–1262 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

