Plasma membrane association of p63 Rho guanine nucleotide exchange factor (p63RhoGEF) is mediated by palmitoylation and is required for basal activity in cells
- PMID: 21832057
- PMCID: PMC3190789
- DOI: 10.1074/jbc.M111.273342
Plasma membrane association of p63 Rho guanine nucleotide exchange factor (p63RhoGEF) is mediated by palmitoylation and is required for basal activity in cells
Abstract
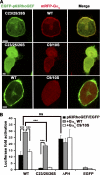
Activation of G protein-coupled receptors at the cell surface leads to the activation or inhibition of intracellular effector enzymes, which include various Rho guanine nucleotide exchange factors (RhoGEFs). RhoGEFs activate small molecular weight GTPases at the plasma membrane (PM). Many of the known G protein-coupled receptor-regulated RhoGEFs are found in the cytoplasm of unstimulated cells, and PM recruitment is a critical aspect of their regulation. In contrast, p63RhoGEF, a Gα(q)-regulated RhoGEF, appears to be constitutively localized to the PM. The objective of this study was to determine the molecular basis for the localization of p63RhoGEF and the impact of its subcellular localization on its regulation by Gα(q). Herein, we show that the pleckstrin homology domain of p63RhoGEF is not involved in its PM targeting. Instead, a conserved string of cysteines (Cys-23/25/26) at the N terminus of the enzyme is palmitoylated and required for membrane localization and full basal activity in cells. Conversion of these residues to serine relocates p63RhoGEF from the PM to the cytoplasm, diminishes its basal activity, and eliminates palmitoylation. The activity of palmitoylation-deficient p63RhoGEF can be rescued by targeting to the PM by fusion with tandem phospholipase C-δ1 pleckstrin homology domains or by co-expression with wild-type Gα(q) but not with palmitoylation-deficient Gα(q). Our data suggest that p63RhoGEF is regulated chiefly through allosteric control by Gα(q), as opposed to other known Gα-regulated RhoGEFs, which are instead sequestered in the cytoplasm, perhaps because of their high basal activity.
Figures





References
-
- Burridge K., Wennerberg K. (2004) Cell 116, 167–179 - PubMed
-
- Heasman S. J., Ridley A. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 690–701 - PubMed
-
- Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 - PubMed
-
- Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 - PubMed
-
- Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

