IL-12 selectively programs effector pathways that are stably expressed in human CD8+ effector memory T cells in vivo
- PMID: 21832277
- PMCID: PMC3193266
- DOI: 10.1182/blood-2011-05-357111
IL-12 selectively programs effector pathways that are stably expressed in human CD8+ effector memory T cells in vivo
Abstract
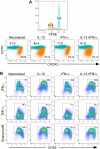
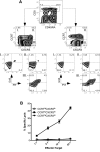
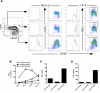
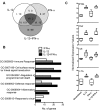
CD8(+) cytotoxic T lymphocytes play a major role in defense against intracellular pathogens, and their functions are specified by antigen recognition and innate cytokines. IL-12 and IFN-α/β are potent "signal 3" cytokines that are involved in both effector and memory cell development. Although the majority of effector cells are eliminated as inflammation resolves, some survive within the pool of memory cells and retain immediate effector function. In this study, we demonstrate that IL-12 instructs a unique program of effector cell differentiation that is distinct from IFN-α/β. Moreover, effector memory (T(EM)) cells within peripheral blood display many common attributes of cells differentiated in vitro in response to IL-12, including proinflammatory cytokine secretion and lytic activity. A pattern of IL-12-induced genes was identified that demarcate T(EM) from central memory cells, and the ontologies of these genes correlated precisely with their effector functions. Further, we uncovered a unique program of gene expression that was acutely regulated by IL-12 and reflected in stable gene expression patterns within T(EM), but not T central memory cells in vivo. Thus, this study directly links a selective set of IL-12-induced genes to the programming of effector functions within the stable population of human CD8(+) T(EM) cells in vivo.
Figures


References
-
- Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. - PubMed
-
- Wherry EJ, Teichgraber V, Becker TC, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4(3):225–234. - PubMed
-
- Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452(7185):356–360. - PubMed
-
- Butler NS, Harty JT. The role of inflammation in the generation and maintenance of memory T cells. Adv Exp Med Biol. 2010;684:42–56. - PubMed
-
- Mescher MF, Curtsinger JM, Agarwal P, et al. Signals required for programming effector and memory development by CD8+ T cells. Immunol Rev. 2006;211:81–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

