Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance
- PMID: 21833332
- PMCID: PMC3153052
- DOI: 10.3389/fmicb.2011.00094
Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance
Abstract
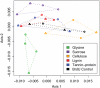
Soils are immensely diverse microbial habitats with thousands of co-existing bacterial, archaeal, and fungal species. Across broad spatial scales, factors such as pH and soil moisture appear to determine the diversity and structure of soil bacterial communities. Within any one site however, bacterial taxon diversity is high and factors maintaining this diversity are poorly resolved. Candidate factors include organic substrate availability and chemical recalcitrance, and given that they appear to structure bacterial communities at the phylum level, we examine whether these factors might structure bacterial communities at finer levels of taxonomic resolution. Analyzing 16S rRNA gene composition of nucleotide analog-labeled DNA by PhyloChip microarrays, we compare relative growth rates on organic substrates of increasing chemical recalcitrance of >2,200 bacterial taxa across 43 divisions/phyla. Taxa that increase in relative abundance with labile organic substrates (i.e., glycine, sucrose) are numerous (>500), phylogenetically clustered, and occur predominantly in two phyla (Proteobacteria and Actinobacteria) including orders Actinomycetales, Enterobacteriales, Burkholderiales, Rhodocyclales, Alteromonadales, and Pseudomonadales. Taxa increasing in relative abundance with more chemically recalcitrant substrates (i.e., cellulose, lignin, or tannin-protein) are fewer (168) but more phylogenetically dispersed, occurring across eight phyla and including Clostridiales, Sphingomonadalaes, Desulfovibrionales. Just over 6% of detected taxa, including many Burkholderiales increase in relative abundance with both labile and chemically recalcitrant substrates. Estimates of median rRNA copy number per genome of responding taxa demonstrate that these patterns are broadly consistent with bacterial growth strategies. Taken together, these data suggest that changes in availability of intrinsically labile substrates may result in predictable shifts in soil bacterial composition.
Keywords: bacteria; bromo-deoxyuridine; carbon; microarray; rRNA copy number; soil; substrate quality.
Figures


References
-
- Allison S. D., Czimczik C. I., Treseder K. K. (2008). Microbial activity and soil respiration under nitrogen addition in alaskan boreal forest. Glob. Change Biol. 14, 1156–1168 10.1111/j.1365-2486.2008.01549.x - DOI
-
- Artursson V., Finlay R. D., Jansson J. K. (2005). Combined bromodeoxyuridine immunocapture and terminal-restriction fragment length polymorphism analysis highlights differences in the active soil bacterial metagenome due to glomus mosseae inoculation or plant species. Environ. Microbiol. 7, 1952–1966 10.1111/j.1462-2920.2005.00868.x - DOI - PubMed
-
- Bader N. E., Cheng W. (2007). Rhizosphere priming effect of populus fremontii obscures the temperature sensitivity of soil organic carbon respiration. Soil Biol. Biochem. 39, 600–606 10.1016/j.soilbio.2006.09.009 - DOI

