Expression of HES and HEY genes in infantile hemangiomas
- PMID: 21834989
- PMCID: PMC3189131
- DOI: 10.1186/2045-824X-3-19
Expression of HES and HEY genes in infantile hemangiomas
Abstract
Background: Infantile hemangiomas (IHs) are the most common benign tumor of infancy, yet their pathogenesis is poorly understood. IHs are believed to originate from a progenitor cell, the hemangioma stem cell (HemSC). Recent studies by our group showed that NOTCH proteins and NOTCH ligands are expressed in hemangiomas, indicating Notch signaling may be active in IHs. We sought to investigate downstream activation of Notch signaling in hemangioma cells by evaluating the expression of the basic HLH family proteins, HES/HEY, in IHs.
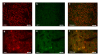
Materials and methods: HemSCs and hemangioma endothelial cells (HemECs) are isolated from freshly resected hemangioma specimens. Quantitative RT-PCR was performed to probe for relative gene transcript levels (normalized to beta-actin). Immunofluorescence was performed to evaluate protein expression. Co-localization studies were performed with CD31 (endothelial cells) and NOTCH3 (peri-vascular, non-endothelial cells). HemSCs were treated with the gamma secretase inhibitor (GSI) Compound E, and gene transcript levels were quantified with real-time PCR.
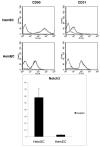
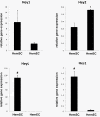
Results: HEY1, HEYL, and HES1 are highly expressed in HemSCs, while HEY2 is highly expressed in HemECs. Protein expression evaluation by immunofluorescence confirms that HEY2 is expressed by HemECs (CD31+ cells), while HEY1, HEYL, and HES1 are more widely expressed and mostly expressed by perivascular cells of hemangiomas. Inhibition of Notch signaling by addition of GSI resulted in down-regulation of HES/HEY genes.
Conclusions: HES/HEY genes are expressed in IHs in cell type specific patterns; HEY2 is expressed in HemECs and HEY1, HEYL, HES1 are expressed in HemSCs. This pattern suggests that HEY/HES genes act downstream of Notch receptors that function in distinct cell types of IHs. HES/HEY gene transcripts are decreased with the addition of a gamma-secretase inhibitor, Compound E, demonstrating that Notch signaling is active in infantile hemangioma cells.
Figures





References
-
- Frieden IJ, Haggstrom AN, Drolet BA, Mancini AJ, Friedlander SF, Boon L, Chamlin SL, Baselga E, Garzon MC, Nopper AJ. et al.Infantile hemangiomas: current knowledge, future directions. Proceedings of a research workshop on infantile hemangiomas, April 7-9, 2005, Bethesda, Maryland, USA. Pediatr Dermatol. 2005;22(5):383–406. doi: 10.1111/j.1525-1470.2005.00102.x. - DOI - PubMed
-
- Alva JA, Iruela-Arispe ML. Notch signaling in vascular morphogenesis. Curr Opin Hematol. 2004;11(4):278–283. doi: 10.1097/01.moh.0000130309.44976.ad. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

