From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem
- PMID: 21835779
- PMCID: PMC3241633
- DOI: 10.1093/nar/gkr573
From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem
Abstract
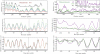
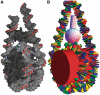
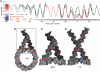
The interaction of histone H1 with linker DNA results in the formation of the nucleosomal stem structure, with considerable influence on chromatin organization. In a recent paper [Syed,S.H., Goutte-Gattat,D., Becker,N., Meyer,S., Shukla,M.S., Hayes,J.J., Everaers,R., Angelov,D., Bednar,J. and Dimitrov,S. (2010) Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl Acad. Sci. USA, 107, 9620-9625], we published results of biochemical footprinting and cryo-electron-micrographs of reconstituted mono-, di- and tri-nucleosomes, for H1 variants with different lengths of the cationic C-terminus. Here, we present a detailed account of the analysis of the experimental data and we include thermal fluctuations into our nano-scale model of the stem structure. By combining (i) crystal and NMR structures of the nucleosome core particle and H1, (ii) the known nano-scale structure and elasticity of DNA, (iii) footprinting information on the location of protected sites on the DNA backbone and (iv) cryo-electron micrographs of reconstituted tri-nucleosomes, we arrive at a description of a polymorphic, hierarchically organized stem with a typical length of 20 ± 2 base pairs. A comparison to linker conformations inferred for poly-601 fibers with different linker lengths suggests, that intra-stem interactions stabilize and facilitate the formation of dense chromatin fibers.
Figures


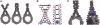



Similar articles
-
Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome.Proc Natl Acad Sci U S A. 2010 May 25;107(21):9620-5. doi: 10.1073/pnas.1000309107. Epub 2010 May 10. Proc Natl Acad Sci U S A. 2010. PMID: 20457934 Free PMC article.
-
Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.Mol Cell. 2017 May 4;66(3):384-397.e8. doi: 10.1016/j.molcel.2017.04.012. Mol Cell. 2017. PMID: 28475873 Free PMC article.
-
The Dynamic Influence of Linker Histone Saturation within the Poly-Nucleosome Array.J Mol Biol. 2021 May 14;433(10):166902. doi: 10.1016/j.jmb.2021.166902. Epub 2021 Mar 2. J Mol Biol. 2021. PMID: 33667509 Free PMC article.
-
The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle.EMBO Rep. 2015 Nov;16(11):1439-53. doi: 10.15252/embr.201540749. Epub 2015 Oct 15. EMBO Rep. 2015. PMID: 26474902 Free PMC article. Review.
-
Histone tail network and modulation in a nucleosome.Curr Opin Struct Biol. 2022 Aug;75:102436. doi: 10.1016/j.sbi.2022.102436. Epub 2022 Jul 18. Curr Opin Struct Biol. 2022. PMID: 35863166 Review.
Cited by
-
Predicting protein-DNA interactions by full search computational docking.Proteins. 2013 Dec;81(12):2106-18. doi: 10.1002/prot.24395. Epub 2013 Oct 18. Proteins. 2013. PMID: 23966176 Free PMC article.
-
Structural insights into the histone H1-nucleosome complex.Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):19390-5. doi: 10.1073/pnas.1314905110. Epub 2013 Nov 11. Proc Natl Acad Sci U S A. 2013. PMID: 24218562 Free PMC article.
-
Unraveling linker histone interactions in nucleosomes.Curr Opin Struct Biol. 2021 Dec;71:87-93. doi: 10.1016/j.sbi.2021.06.001. Epub 2021 Jul 8. Curr Opin Struct Biol. 2021. PMID: 34246862 Free PMC article. Review.
-
Toward convergence of experimental studies and theoretical modeling of the chromatin fiber.J Biol Chem. 2012 Feb 17;287(8):5183-91. doi: 10.1074/jbc.R111.305763. Epub 2011 Dec 7. J Biol Chem. 2012. PMID: 22157002 Free PMC article. Review.
-
Linker histones: novel insights into structure-specific recognition of the nucleosome.Biochem Cell Biol. 2017 Apr;95(2):171-178. doi: 10.1139/bcb-2016-0097. Epub 2016 Jun 29. Biochem Cell Biol. 2017. PMID: 28177778 Free PMC article. Review.
References
-
- Ball P. Portrait of a molecule. Nature. 2003;421:421–422. - PubMed
-
- Kornberg RD. Structure of chromatin. Annu. Rev. Biochem. 1997;46:931–954. 1977. - PubMed
-
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Van Holde KE. Chromatin. New York: Springer; 1989.
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature. 1997;389:251–260. - PubMed

