Conformation-selective methylation of geminivirus DNA
- PMID: 21835804
- PMCID: PMC3209285
- DOI: 10.1128/JVI.05567-11
Conformation-selective methylation of geminivirus DNA
Abstract
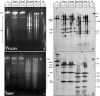
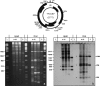
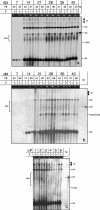
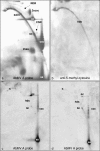
Geminiviruses with small circular single-stranded DNA genomes replicate in plant cell nuclei by using various double-stranded DNA (dsDNA) intermediates: distinct open circular and covalently closed circular as well as heterogeneous linear DNA. Their DNA may be methylated partially at cytosine residues, as detected previously by bisulfite sequencing and subsequent PCR. In order to determine the methylation patterns of the circular molecules, the DNAs of tomato yellow leaf curl Sardinia virus (TYLCSV) and Abutilon mosaic virus were investigated utilizing bisulfite treatment followed by rolling circle amplification. Shotgun sequencing of the products yielded a randomly distributed 50% rate of C maintenance after the bisulfite reaction for both viruses. However, controls with unmethylated single-stranded bacteriophage DNA resulted in the same level of C maintenance. Only one short DNA stretch within the C2/C3 promoter of TYLCSV showed hyperprotection of C, with the protection rate exceeding the threshold of the mean value plus 1 standard deviation. Similarly, the use of methylation-sensitive restriction enzymes suggested that geminiviruses escape silencing by methylation very efficiently, by either a rolling circle or recombination-dependent replication mode. In contrast, attempts to detect methylated bases positively by using methylcytosine-specific antibodies detected methylated DNA only in heterogeneous linear dsDNA, and methylation-dependent restriction enzymes revealed that the viral heterogeneous linear dsDNA was methylated preferentially.
Figures


References
-
- Abouzid A. M., Frischmuth T., Jeske H. 1988. A putative replicative form of the abutilon mosaic virus (gemini group) in a chromatin-like structure. Mol. Gen. Genet. 212: 252–258
-
- Alberter B., Rezaian A. M., Jeske H. 2005. Replicative intermediates of ToLCV and its satellite DNAs. Virology 331: 441–448 - PubMed
-
- Bian X.-Y., Rasheed M. S., Seemanpillai M., Rezaian A. 2006. Analysis of silencing escape of tomato leaf curl virus: an evaluation of the role of DNA methylation. Mol. Plant Microbe Interact. 19: 614–624 - PubMed
-
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

