Inference for discretely observed stochastic kinetic networks with applications to epidemic modeling
- PMID: 21835814
- PMCID: PMC3276272
- DOI: 10.1093/biostatistics/kxr019
Inference for discretely observed stochastic kinetic networks with applications to epidemic modeling
Abstract
We present a new method for Bayesian Markov Chain Monte Carlo-based inference in certain types of stochastic models, suitable for modeling noisy epidemic data. We apply the so-called uniformization representation of a Markov process, in order to efficiently generate appropriate conditional distributions in the Gibbs sampler algorithm. The approach is shown to work well in various data-poor settings, that is, when only partial information about the epidemic process is available, as illustrated on the synthetic data from SIR-type epidemics and the Center for Disease Control and Prevention data from the onset of the H1N1 pandemic in the United States.
Figures
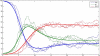

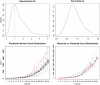
Similar articles
-
Capturing the time-varying drivers of an epidemic using stochastic dynamical systems.Biostatistics. 2013 Jul;14(3):541-55. doi: 10.1093/biostatistics/kxs052. Epub 2013 Jan 4. Biostatistics. 2013. PMID: 23292757
-
Bayesian inference for stochastic multitype epidemics in structured populations using sample data.Biostatistics. 2009 Oct;10(4):779-91. doi: 10.1093/biostatistics/kxp031. Epub 2009 Jul 31. Biostatistics. 2009. PMID: 19648227
-
Bayesian model choice for epidemic models with two levels of mixing.Biostatistics. 2014 Jan;15(1):46-59. doi: 10.1093/biostatistics/kxt023. Epub 2013 Jul 24. Biostatistics. 2014. PMID: 23887980
-
Modelling under-reporting in epidemics.J Math Biol. 2014 Sep;69(3):737-65. doi: 10.1007/s00285-013-0717-z. Epub 2013 Aug 13. J Math Biol. 2014. PMID: 23942791
-
Phylogenetic and epidemic modeling of rapidly evolving infectious diseases.Infect Genet Evol. 2011 Dec;11(8):1825-41. doi: 10.1016/j.meegid.2011.08.005. Epub 2011 Aug 31. Infect Genet Evol. 2011. PMID: 21906695 Free PMC article. Review.
Cited by
-
Network reconstruction from infection cascades.J R Soc Interface. 2019 Feb 28;16(151):20180844. doi: 10.1098/rsif.2018.0844. J R Soc Interface. 2019. PMID: 30958195 Free PMC article.
-
The multilevel hierarchical data EM-algorithm. Applications to discrete-time Markov chain epidemic models.Heliyon. 2022 Dec 22;8(12):e12622. doi: 10.1016/j.heliyon.2022.e12622. eCollection 2022 Dec. Heliyon. 2022. PMID: 36643325 Free PMC article.
-
Comparison of Parameter Estimation Methods in Stochastic Chemical Kinetic Models: Examples in Systems Biology.AIChE J. 2014 Apr;60(4):1253-1268. doi: 10.1002/aic.14409. Epub 2014 Mar 5. AIChE J. 2014. PMID: 27429455 Free PMC article.
-
Survival dynamical systems: individual-level survival analysis from population-level epidemic models.Interface Focus. 2020 Feb 6;10(1):20190048. doi: 10.1098/rsfs.2019.0048. Epub 2019 Dec 13. Interface Focus. 2020. PMID: 31897290 Free PMC article.
-
COVID-19: Estimation of the transmission dynamics in Spain using a stochastic simulator and black-box optimization techniques.Comput Methods Programs Biomed. 2021 Nov;211:106399. doi: 10.1016/j.cmpb.2021.106399. Epub 2021 Sep 6. Comput Methods Programs Biomed. 2021. PMID: 34607036 Free PMC article.
References
-
- Andersson H, Britton T. Stochastic Epidemic Models and Their Statistical Analysis. New York: Springer; 2000a.
-
- Andersson H, Britton T. Stochastic epidemics in dynamic populations: quasi-stationarity and extinction. Journal of Mathematical Biology. 2000b;41:559–580. - PubMed
-
- Bellman R, Dreyfus S. Functional Approximations and Dynamic Programming. Mathematical Tables and Other Aids to Computation. 1959;13:247–251.
-
- Boys R, Giles P. Bayesian inference for stochastic epidemic models with time-inhomogeneous removal rates. Journal of Mathematical Biology. 2007;55:223–247. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

