A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial
- PMID: 21836613
- PMCID: PMC4545515
- DOI: 10.1038/leu.2011.204
A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial
Abstract
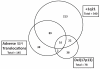
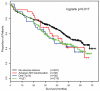
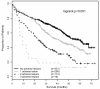
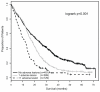
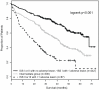
The association of genetic lesions detected by fluorescence in situ hybridization (FISH) with survival was analyzed in 1069 patients with newly presenting myeloma treated in the Medical Research Council Myeloma IX trial, with the aim of identifying patients associated with the worst prognosis. A comprehensive FISH panel was performed, and the lesions associated with short progression-free survival and overall survival (OS) in multivariate analysis were +1q21, del(17p13) and an adverse immunoglobulin heavy chain gene (IGH) translocation group incorporating t(4;14), t(14;16) and t(14;20). These lesions frequently co-segregated, and there was an association between the accumulation of these adverse FISH lesions and a progressive impairment of survival. This observation was used to define a series of risk groups based on number of adverse lesions. Taking this approach, we defined a favorable risk group by the absence of adverse genetic lesions, an intermediate group with one adverse lesion and a high-risk group defined by the co-segregation of >1 adverse lesion. This genetic grouping was independent of the International Staging System (ISS) and so was integrated with the ISS to identify an ultra-high-risk group defined by ISS II or III and >1 adverse lesion. This group constituted 13.8% of patients and was associated with a median OS of 19.4 months.
Figures

References
-
- Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005 May 20;23(15):3412–3420. - PubMed
-
- Shaughnessy JD, Jr., Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007 Mar 15;109(6):2276–2284. - PubMed
-
- DecauxO LL, Magrangeas F, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiplod signatures in low risk patients: A study of the Intergroupe Francophone dy Myelome. J Clin Oncol. 2008;26(24) - PubMed
-
- Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003 Feb 15;101(4):1520–1529. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

