The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators
- PMID: 21836637
- PMCID: PMC3185338
- DOI: 10.1038/embor.2011.152
The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators
Abstract
Covalent attachment of small ubiquitin-like modifier (SUMO) to proteins regulates many processes in the eukaryotic cell. This reaction is similar to ubiquitination and usually requires an E3 ligase for substrate modification. However, only a few SUMO ligases have been described so far, which frequently facilitate sumoylation by bringing together the SUMO-conjugating enzyme Ubc9 and the target protein. Ubc9 is an interaction partner of the transcription factor Krox20, a key regulator of hindbrain development. Here, we show that Krox20 functions as a SUMO ligase for its coregulators--the Nab proteins--and that Nab sumoylation negatively modulates Krox20 transcriptional activity in vivo.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
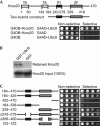


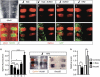
References
-
- Chomette D, Frain M, Cereghini S, Charnay P, Ghislain J (2006) Krox20 hindbrain cis-regulatory landscape: interplay between multiple long-range initiation and autoregulatory elements. Development 133: 1253–1262 - PubMed
-
- Garcia-Dominguez M, Reyes JC (2009) SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789: 451–459 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

