Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in India
- PMID: 21837798
- PMCID: PMC4412166
- DOI: 10.1002/jmv.22176
Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in India
Abstract
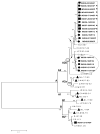
Human metapneumovirus (hMPV) causes acute respiratory infections in children and adults. It is classified into two major genetic lineages and each lineage into two sublineages. The purpose of the study was to identify and characterize hMPV in children who presented to the All India Institute of Medical Sciences, New Delhi, India with acute respiratory infection from April 2005 to March 2007. By reverse-transcription polymerase chain reaction, hMPV was detected in 21 (3%) of the 662 nasopharyngeal samples from children with acute respiratory infection and in none of the 120 control children. Seven of the 21 (33%) children infected with hMPV required hospital admission for pneumonia or bronchiolitis. Most hMPV detections were during the winter and spring seasons. The majority (67%, 11/21) of children positive for hMPV were within 24 months of age. Phylogenetic analysis of partial F and N gene and the full G gene sequences showed three sub-lineages of hMPV circulated during the study period, B1, B2, and the novel sub-lineage A2b. The circulation pattern of hMPV genotypes varied by season. Comparison of the F and G genes of eight strains revealed incongruencies in lineage assignments, raising the possibility that recombination had occurred. Sequence analysis also revealed the F gene was relatively conserved whereas the G gene was more variable between the A and B lineages. This study demonstrates that hMPV is an important contributor to acute respiratory infection in children in India, resulting in both outpatient visits and hospitalizations.
Copyright © 2011 Wiley-Liss, Inc.
Figures





References
-
- Agapov E, Sumino KC, Gaudreault-Keener M, Storch GA, Holtzman MJ. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J Infect Dis. 2006;193:396–403. - PubMed
-
- Banerjee S, Bharaj P, Sullender W, Kabra SK, Broor S. Human metapneumovirus infections among children with acute respiratory infections seen in a large referral hospital in India. J Clin Virol. 2007;38:70–72. - PubMed
-
- Bharaj P, Sullender WM, Kabra SK, Mani K, Cherian J, Tyagi V, Chahar HS, Kaushik S, Dar L, Broor S. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89. - PMC - PubMed
-
- Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315:1–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

