Molecular imaging with nucleic acid aptamers
- PMID: 21838686
- PMCID: PMC3205285
- DOI: 10.2174/092986711797189691
Molecular imaging with nucleic acid aptamers
Abstract
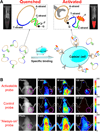
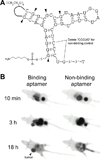
With many desirable properties such as ease of synthesis, small size, lack of immunogenicity, and versatile chemistry, aptamers represent a class of targeting ligands that possess tremendous potential in molecular imaging applications. Non-invasive imaging of various disease markers with aptamer-based probes has many potential clinical applications such as lesion detection, patient stratification, treatment monitoring, etc. In this review, we will summarize the current status of molecular imaging with aptamer-based probes. First, fluorescence imaging will be described which include both direct targeting and activatable probes. Next, we discuss molecular magnetic resonance imaging and targeted ultrasound investigations using aptamer-based agents. Radionuclide-based imaging techniques (single-photon emission computed tomography and positron emission tomography) will be summarized as well. In addition, aptamers have also been labeled with various tags for computed tomography, surface plasmon resonance, dark-field light scattering microscopy, transmission electron microscopy, and surface-enhanced Raman spectroscopy imaging. Among all molecular imaging modalities, no single modality is perfect and sufficient to obtain all the necessary information for a particular question. Thus, a multimodality probe has also been constructed for concurrent fluorescence, gamma camera, and magnetic resonance imaging in vivo. Although the future of aptamer-based molecular imaging is becoming increasingly bright and many proof-of-principle studies have already been reported, much future effort needs to be directed towards the development of clinically translatable aptamer-based imaging agents which will eventually benefit patients.
Figures




Similar articles
-
Aptamers used for molecular imaging and theranostics - recent developments.Theranostics. 2022 May 13;12(9):4010-4050. doi: 10.7150/thno.72949. eCollection 2022. Theranostics. 2022. PMID: 35673581 Free PMC article. Review.
-
Applications of aptamers in targeted imaging: state of the art.Curr Top Med Chem. 2015;15(12):1138-52. doi: 10.2174/1568026615666150413153400. Curr Top Med Chem. 2015. PMID: 25866268 Free PMC article. Review.
-
Application of aptamers for in vivo molecular imaging and theranostics.Adv Drug Deliv Rev. 2018 Sep;134:94-106. doi: 10.1016/j.addr.2018.08.004. Epub 2018 Aug 17. Adv Drug Deliv Rev. 2018. PMID: 30125606 Review.
-
Current progress of aptamer-based molecular imaging.J Nucl Med. 2014 Mar;55(3):353-6. doi: 10.2967/jnumed.113.126144. Epub 2014 Feb 13. J Nucl Med. 2014. PMID: 24525205 Free PMC article. Review.
-
Aptamers: versatile molecular recognition probes for cancer detection.Analyst. 2016 Jan 21;141(2):403-15. doi: 10.1039/c5an01995h. Analyst. 2016. PMID: 26618445 Free PMC article. Review.
Cited by
-
Image guided biodistribution and pharmacokinetic studies of theranostics.Theranostics. 2012;2(11):1040-53. doi: 10.7150/thno.4652. Epub 2012 Nov 5. Theranostics. 2012. PMID: 23227121 Free PMC article. Review.
-
The Potential of Aptamer-Mediated Liquid Biopsy for Early Detection of Cancer.Int J Mol Sci. 2021 May 25;22(11):5601. doi: 10.3390/ijms22115601. Int J Mol Sci. 2021. PMID: 34070509 Free PMC article. Review.
-
Aptamers Selected by Cell-SELEX for Molecular Imaging.J Mol Evol. 2015 Dec;81(5-6):162-71. doi: 10.1007/s00239-015-9716-6. Epub 2015 Nov 19. J Mol Evol. 2015. PMID: 26584804 Free PMC article. Review.
-
Tetraspanins as regulators of the tumour microenvironment: implications for metastasis and therapeutic strategies.Br J Pharmacol. 2014 Dec;171(24):5462-90. doi: 10.1111/bph.12260. Br J Pharmacol. 2014. PMID: 23731188 Free PMC article. Review.
-
Novel Aptamer Strategies in Combating Bacterial Infections: From Diagnostics to Therapeutics.Pharmaceutics. 2024 Aug 29;16(9):1140. doi: 10.3390/pharmaceutics16091140. Pharmaceutics. 2024. PMID: 39339177 Free PMC article. Review.
References
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. - PubMed
-
- Takafuji Y, Jo JI, Tabata Y. Simple PEG modification of DNA aptamer based on copper ion coordination for tumor targeting. J. Biomater. Sci. Polym. Ed. 2010 Epub. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

