A statistical framework for eQTL mapping using RNA-seq data
- PMID: 21838806
- PMCID: PMC3218220
- DOI: 10.1111/j.1541-0420.2011.01654.x
A statistical framework for eQTL mapping using RNA-seq data
Abstract
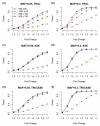
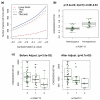
RNA-seq may replace gene expression microarrays in the near future. Using RNA-seq, the expression of a gene can be estimated using the total number of sequence reads mapped to that gene, known as the total read count (TReC). Traditional expression quantitative trait locus (eQTL) mapping methods, such as linear regression, can be applied to TReC measurements after they are properly normalized. In this article, we show that eQTL mapping, by directly modeling TReC using discrete distributions, has higher statistical power than the two-step approach: data normalization followed by linear regression. In addition, RNA-seq provides information on allele-specific expression (ASE) that is not available from microarrays. By combining the information from TReC and ASE, we can computationally distinguish cis- and trans-eQTL and further improve the power of cis-eQTL mapping. Both simulation and real data studies confirm the improved power of our new methods. We also discuss the design issues of RNA-seq experiments. Specifically, we show that by combining TReC and ASE measurements, it is possible to minimize cost and retain the statistical power of cis-eQTL mapping by reducing sample size while increasing the number of sequence reads per sample. In addition to RNA-seq data, our method can also be employed to study the genetic basis of other types of sequencing data, such as chromatin immunoprecipitation followed by DNA sequencing data. In this article, we focus on eQTL mapping of a single gene using the association-based method. However, our method establishes a statistical framework for future developments of eQTL mapping methods using RNA-seq data (e.g., linkage-based eQTL mapping), and the joint study of multiple genetic markers and/or multiple genes.
© 2011, The International Biometric Society.
Figures


References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300.
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
-
- Byrd R, Lu P, Nocedal J, Zhu C. A limited memory algorithm for bound constrained optimization. SIAM Journal on Scientific Computing. 1995;16:1190–1208.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

