Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma
- PMID: 21840481
- PMCID: PMC4154494
- DOI: 10.1016/j.ccr.2011.07.007
Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma
Abstract
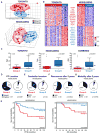
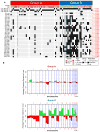
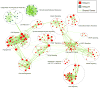
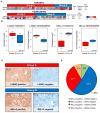
Despite the histological similarity of ependymomas from throughout the neuroaxis, the disease likely comprises multiple independent entities, each with a distinct molecular pathogenesis. Transcriptional profiling of two large independent cohorts of ependymoma reveals the existence of two demographically, transcriptionally, genetically, and clinically distinct groups of posterior fossa (PF) ependymomas. Group A patients are younger, have laterally located tumors with a balanced genome, and are much more likely to exhibit recurrence, metastasis at recurrence, and death compared with Group B patients. Identification and optimization of immunohistochemical (IHC) markers for PF ependymoma subgroups allowed validation of our findings on a third independent cohort, using a human ependymoma tissue microarray, and provides a tool for prospective prognostication and stratification of PF ependymoma patients.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Posterior fossa ependymomas: a tale of two subtypes.Cancer Cell. 2011 Aug 16;20(2):133-4. doi: 10.1016/j.ccr.2011.08.003. Cancer Cell. 2011. PMID: 21840477
References
-
- Bouffet E, Perilongo G, Canete A, Massimino M. Intracranial ependymomas in children: a critical review of prognostic factors and a plea for cooperation. Med Pediatr Oncol. 1998;30:319–329. discussion 329–331. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

