The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB
- PMID: 21840978
- PMCID: PMC3187222
- DOI: 10.1128/JB.05424-11
The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB
Abstract
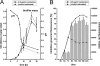
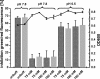
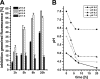
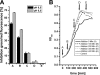
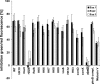
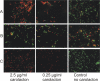
Carolacton, a secondary metabolite isolated from the myxobacterium Sorangium cellulosum, disturbs Streptococcus mutans biofilm viability at nanomolar concentrations. Here we show that carolacton causes leakage of cytoplasmic content (DNA and proteins) in growing cells at low pH and provide quantitative data on the membrane damage. Furthermore, we demonstrate that the biofilm-specific activity of carolacton is due to the strong acidification occurring during biofilm growth. The chemical conversion of the ketocarbonic function of the molecule to a carolacton methylester did not impact its activity, indicating that carolacton is not functionally activated at low pH by a change of its net charge. A comparative time series microarray analysis identified the VicKRX and ComDE two-component signal transduction systems and genes involved in cell wall metabolism as playing essential roles in the response to carolacton treatment. A sensitivity testing of mutants with deletions of all 13 viable histidine kinases and the serine/threonine protein kinase PknB of S. mutans identified only the ΔpknB deletion mutant as being insensitive to carolacton treatment. A strong overlap between the regulon of PknB in S. mutans and the genes affected by carolacton treatment was found. The data suggest that carolacton acts by interfering with PknB-mediated signaling in growing cells. The resulting altered cell wall morphology causes membrane damage and cell death at low pH.
Figures
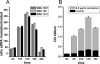





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

