miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans
- PMID: 21841313
- PMCID: PMC3163949
- DOI: 10.1172/JCI45390
miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans
Erratum in
-
miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans.J Clin Invest. 2017 Mar 1;127(3):1116. doi: 10.1172/JCI92099. Epub 2017 Mar 1. J Clin Invest. 2017. PMID: 28248205 Free PMC article. No abstract available.
Abstract
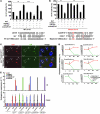
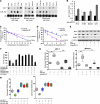
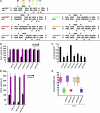
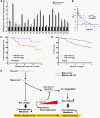
MicroRNAs (miRNAs) influence many biological processes, including cancer. They do so by posttranscriptionally repressing target mRNAs to which they have sequence complementarity. Although it has been postulated that miRNAs can regulate other miRNAs, this has never been shown experimentally to our knowledge. Here, we demonstrate that miR-107 negatively regulates the tumor suppressor miRNA let-7 via a direct interaction. miR-107 was found to be highly expressed in malignant tissue from patients with advanced breast cancer, and its expression was inversely correlated with let-7 expression in tumors and in cancer cell lines. Ectopic expression of miR-107 in human cancer cell lines led to destabilization of mature let-7, increased expression of let-7 targets, and increased malignant phenotypes. In contrast, depletion of endogenous miR-107 dramatically increased the stability of mature let-7 and led to downregulation of let-7 targets. Accordingly, miR-107 expression increased the tumorigenic and metastatic potential of a human breast cancer cell line in mice via inhibition of let-7 and upregulation of let-7 targets. By mutating individual sites within miR-107 and let-7, we found that miR-107 directly interacts with let-7 and that the internal loop of the let-7/miR-107 duplex is critical for repression of let-7 expression. Altogether, we have identified an oncogenic role for miR-107 and provide evidence of a transregulational interaction among miRNAs in human cancer development.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

