Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation
- PMID: 21841785
- PMCID: PMC3669688
- DOI: 10.1038/ni.2084
Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation
Abstract
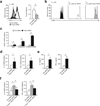
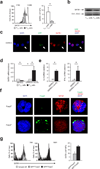
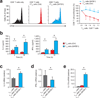
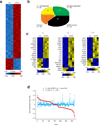
Regulatory T cells (T(reg) cells) are essential for self-tolerance and immune homeostasis. Lack of effector T cell (T(eff) cell) function and gain of suppressive activity by T(reg) cells are dependent on the transcriptional program induced by Foxp3. Here we report that repression of SATB1, a genome organizer that regulates chromatin structure and gene expression, was crucial for the phenotype and function of T(reg) cells. Foxp3, acting as a transcriptional repressor, directly suppressed the SATB1 locus and indirectly suppressed it through the induction of microRNAs that bound the SATB1 3' untranslated region. Release of SATB1 from the control of Foxp3 in T(reg) cells caused loss of suppressive function, establishment of transcriptional T(eff) cell programs and induction of T(eff) cell cytokines. Our data support the proposal that inhibition of SATB1-mediated modulation of global chromatin remodeling is pivotal for maintaining T(reg) cell functionality.
Figures




References
-
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. - PubMed
-
- Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. - PubMed
-
- Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. - PubMed
-
- Lahl K, et al. Nonfunctional regulatory T cells and defective control of Th2 cytokine production in natural scurfy mutant mice. J Immunol. 2009;183:5662–5672. - PubMed
-
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

