The Cryptochrome Blue Light Receptors
- PMID: 21841916
- PMCID: PMC3155252
- DOI: 10.1199/tab.0135
The Cryptochrome Blue Light Receptors
Abstract
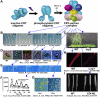
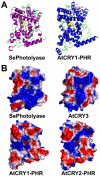
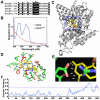
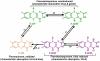
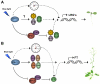
Cryptochromes are photolyase-like blue light receptors originally discovered in Arabidopsis but later found in other plants, microbes, and animals. Arabidopsis has two cryptochromes, CRY1 and CRY2, which mediate primarily blue light inhibition of hypocotyl elongation and photoperiodic control of floral initiation, respectively. In addition, cryptochromes also regulate over a dozen other light responses, including circadian rhythms, tropic growth, stomata opening, guard cell development, root development, bacterial and viral pathogen responses, abiotic stress responses, cell cycles, programmed cell death, apical dominance, fruit and ovule development, seed dormancy, and magnetoreception. Cryptochromes have two domains, the N-terminal PHR (Photolyase-Homologous Region) domain that bind the chromophore FAD (flavin adenine dinucleotide), and the CCE (CRY C-terminal Extension) domain that appears intrinsically unstructured but critical to the function and regulation of cryptochromes. Most cryptochromes accumulate in the nucleus, and they undergo blue light-dependent phosphorylation or ubiquitination. It is hypothesized that photons excite electrons of the flavin molecule, resulting in redox reaction or circular electron shuttle and conformational changes of the photoreceptors. The photoexcited cryptochrome are phosphorylated to adopt an open conformation, which interacts with signaling partner proteins to alter gene expression at both transcriptional and posttranslational levels and consequently the metabolic and developmental programs of plants.
Figures
References
-
- Ahmad M., Cashmore A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;3668(1):162–166. - PubMed
-
- Ahmad M., Lin C., Cashmore A.R. Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 1995;88(1):653–658. - PubMed
-
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature. 1998b;3928(1):720–723. - PubMed
-
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998c;18(1):939–948. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
