Diffusion models of protein folding
- PMID: 21842082
- PMCID: PMC3457642
- DOI: 10.1039/c1cp21541h
Diffusion models of protein folding
Abstract
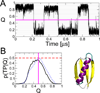
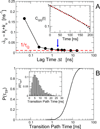
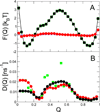
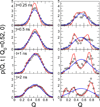
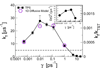
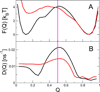
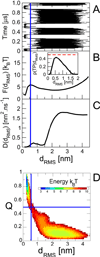
In theory and in the analysis of experiments, protein folding is often described as diffusion along a single coordinate. We explore here the application of a one-dimensional diffusion model to interpret simulations of protein folding, where the parameters of a model that "best" describes the simulation trajectories are determined using a Bayesian analysis. We discuss the requirements for such a model to be a good approximation to the global dynamics, and several methods for testing its accuracy. For example, one test considers the effect of an added bias potential on the fitted free energies and diffusion coefficients. Such a bias may also be used to extend our approach to determining parameters for the model to systems that would not normally explore the full coordinate range on accessible time scales. Alternatively, the propagators predicted from the model at different "lag" times may be compared with observations from simulation. We then present some applications of the model to protein folding, including Kramers-like turnover in folding rates of coarse-grained models, the effect of non-native interactions on folding, and the effect of the chosen coordinate on the observed position-dependence of the diffusion coefficients. Lastly, we consider how our results are useful for the interpretation of experiments, and how this type of Bayesian analysis may eventually be applied directly to analyse experimental data.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

