Facile assembly of micro- and nanoarrays for sensing with natural cell membranes
- PMID: 21842844
- PMCID: PMC3183111
- DOI: 10.1021/nn202554t
Facile assembly of micro- and nanoarrays for sensing with natural cell membranes
Abstract
Microarray technology has facilitated many powerful high-throughput studies in the fields of genetics and proteomics, among others. However, preparation of microarrays composed of cell-derived membranes with embedded receptors has proven difficult. Here we describe a new method for forming microarrays composed of synthetic lipid vesicles and natural cell membranes. The method is based upon assembly of vesicles and natural membranes into recessed micro- and nanowells and using a polydimethylsiloxane (PDMS) block as a "squeegee." This method is used to assemble phospholipid vesicles into arrays with micrometer and nanoscale dimensions. Native myelin and neuronal lipid raft arrays are also formed in 30 min or less. We show the natural membrane arrays can be used for sensing lipid-protein interactions by detecting cholera toxin binding to ganglioside GM1 in neuronal lipid rafts. In multicomponent arrays myelin can be distinguished from neuronal rafts by antibody binding to cell-specific surface antigens. Finally, myelin arrays formed in gold nanowells are used for surface plasmon resonance sensing. This assembly approach is simple, broadly applicable, and opens up new avenues of research not easily accomplished with standard microarray technology.
© 2011 American Chemical Society
Figures

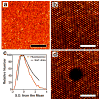
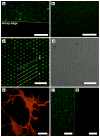

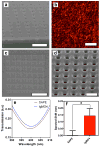
References
-
- MacBeath G, Schreiber SL. Printing Proteins as Microarrays for High-throughput Function Determination. Science. 2000;289:1760–1763. - PubMed
-
- Kodadek T. Protein Microarrays: Prospects and Problems. Chem Biol. 2001;8:105–115. - PubMed
-
- Feizi T, Fazio F, Chai W, Wong C. Carbohydrate Microarrays - A New Set of Technologies at the Frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. - PubMed
-
- Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling Protein Microarrays. Science. 2004;305:86–90. - PubMed
-
- Jones RB, Gordus A, Krall JA, MacBeath G. A Quantitative Protein Interaction Network for the ErbB Receptors Using Protein Microarrays. Nature. 2006;439:168–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

