Understanding the errors of SHAPE-directed RNA structure modeling
- PMID: 21842868
- PMCID: PMC3172344
- DOI: 10.1021/bi200524n
Understanding the errors of SHAPE-directed RNA structure modeling
Abstract
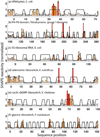
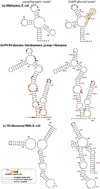
Single-nucleotide-resolution chemical mapping for structured RNA is being rapidly advanced by new chemistries, faster readouts, and coupling to computational algorithms. Recent tests have shown that selective 2'-hydroxyl acylation by primer extension (SHAPE) can give near-zero error rates (0-2%) in modeling the helices of RNA secondary structure. Here, we benchmark the method using six molecules for which crystallographic data are available: tRNA(phe) and 5S rRNA from Escherichia coli, the P4-P6 domain of the Tetrahymena group I ribozyme, and ligand-bound domains from riboswitches for adenine, cyclic di-GMP, and glycine. SHAPE-directed modeling of these highly structured RNAs gave an overall false negative rate (FNR) of 17% and a false discovery rate (FDR) of 21%, with at least one helix prediction error in five of the six cases. Extensive variations of data processing, normalization, and modeling parameters did not significantly mitigate modeling errors. Only one varation, filtering out data collected with deoxyinosine triphosphate during primer extension, gave a modest improvement (FNR = 12%, and FDR = 14%). The residual structure modeling errors are explained by the insufficient information content of these RNAs' SHAPE data, as evaluated by a nonparametric bootstrapping analysis. Beyond these benchmark cases, bootstrapping suggests a low level of confidence (<50%) in the majority of helices in a previously proposed SHAPE-directed model for the HIV-1 RNA genome. Thus, SHAPE-directed RNA modeling is not always unambiguous, and helix-by-helix confidence estimates, as described herein, may be critical for interpreting results from this powerful methodology.
© 2011 American Chemical Society
Figures

Comment in
-
Statistical analysis of SHAPE-directed RNA secondary structure modeling.Biochemistry. 2013 Jan 29;52(4):596-9. doi: 10.1021/bi300756s. Epub 2013 Jan 14. Biochemistry. 2013. PMID: 23286327 Free PMC article.
Similar articles
-
Principles for understanding the accuracy of SHAPE-directed RNA structure modeling.Biochemistry. 2013 Jan 29;52(4):588-95. doi: 10.1021/bi300755u. Epub 2013 Jan 14. Biochemistry. 2013. PMID: 23316814 Free PMC article.
-
Statistical analysis of SHAPE-directed RNA secondary structure modeling.Biochemistry. 2013 Jan 29;52(4):596-9. doi: 10.1021/bi300756s. Epub 2013 Jan 14. Biochemistry. 2013. PMID: 23286327 Free PMC article.
-
The 3D arrangement of the 23 S and 5 S rRNA in the Escherichia coli 50 S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 A resolution.J Mol Biol. 2000 Apr 21;298(1):35-59. doi: 10.1006/jmbi.2000.3635. J Mol Biol. 2000. PMID: 10756104
-
The nucleotide sequence of 5 S ribosomal RNA from Vibrio marinus.Microbiol Sci. 1984 Dec;1(9):229-31. Microbiol Sci. 1984. PMID: 6086080 Review.
-
Structure and functions of 5S rRNA.Acta Biochim Pol. 2001;48(1):191-8. Acta Biochim Pol. 2001. PMID: 11440169 Review.
Cited by
-
Database proton NMR chemical shifts for RNA signal assignment and validation.J Biomol NMR. 2013 Jan;55(1):33-46. doi: 10.1007/s10858-012-9683-9. Epub 2012 Nov 23. J Biomol NMR. 2013. PMID: 23180050 Free PMC article.
-
RNA design rules from a massive open laboratory.Proc Natl Acad Sci U S A. 2014 Feb 11;111(6):2122-7. doi: 10.1073/pnas.1313039111. Epub 2014 Jan 27. Proc Natl Acad Sci U S A. 2014. PMID: 24469816 Free PMC article.
-
The centrality of RNA for engineering gene expression.Biotechnol J. 2013 Dec;8(12):1379-95. doi: 10.1002/biot.201300018. Epub 2013 Oct 4. Biotechnol J. 2013. PMID: 24124015 Free PMC article. Review.
-
Consistent features observed in structural probing data of eukaryotic RNAs.NAR Genom Bioinform. 2025 Jan 30;7(1):lqaf001. doi: 10.1093/nargab/lqaf001. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 39885881 Free PMC article.
-
RNA structure: Adding a second dimension.Nat Chem. 2011 Nov 23;3(12):913-5. doi: 10.1038/nchem.1209. Nat Chem. 2011. PMID: 22109268 No abstract available.
References
-
- Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. - PubMed
-
- Gesteland RF, Cech TR, Atkins JF. The RNA world: the nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006.
-
- Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

