Amplification, mutation, and sequencing of a six-letter synthetic genetic system
- PMID: 21842904
- PMCID: PMC3427765
- DOI: 10.1021/ja204910n
Amplification, mutation, and sequencing of a six-letter synthetic genetic system
Abstract
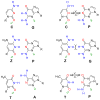
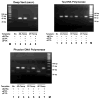
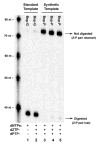
The next goals in the development of a synthetic biology that uses artificial genetic systems will require chemistry-biology combinations that allow the amplification of DNA containing any number of sequential and nonsequential nonstandard nucleotides. This amplification must ensure that the nonstandard nucleotides are not unidirectionally lost during PCR amplification (unidirectional loss would cause the artificial system to revert to an all-natural genetic system). Further, technology is needed to sequence artificial genetic DNA molecules. The work reported here meets all three of these goals for a six-letter artificially expanded genetic information system (AEGIS) that comprises four standard nucleotides (G, A, C, and T) and two additional nonstandard nucleotides (Z and P). We report polymerases and PCR conditions that amplify a wide range of GACTZP DNA sequences having multiple consecutive unnatural synthetic genetic components with low (0.2% per theoretical cycle) levels of mutation. We demonstrate that residual mutation processes both introduce and remove unnatural nucleotides, allowing the artificial genetic system to evolve as such, rather than revert to a wholly natural system. We then show that mechanisms for these residual mutation processes can be exploited in a strategy to sequence "six-letter" GACTZP DNA. These are all not yet reported for any other synthetic genetic system.
Figures





References
-
- Szybalski W. In vivo and in vitro Initiation of transcription. In: Kohn A, Shatkay A, editors. Control of Gene Expression. New York: Plenum Press; 1974. pp. 23–24.pp. 404–405.pp. 411–412.pp. 415–417.
-
- Henry AA, Romesberg FE. Current Opin Chem Biol. 2003;7:727–733. - PubMed
-
- Piccirilli JA, Krauch T, Moroney SE, Benner SA. Nature. 1990;343:33–37. - PubMed
-
- Bain JD, Chamberlin AR, Switzer CY, Benner SA. Nature. 1992;356:537–539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

