Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models
- PMID: 21843480
- PMCID: PMC3175065
- DOI: 10.1016/j.bpj.2011.06.046
Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models
Abstract
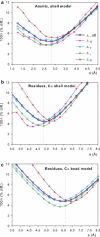
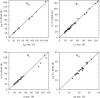
Here we extend the ability to predict hydrodynamic coefficients and other solution properties of rigid macromolecular structures from atomic-level structures, implemented in the computer program HYDROPRO, to models with lower, residue-level resolution. Whereas in the former case there is one bead per nonhydrogen atom, the latter contains one bead per amino acid (or nucleotide) residue, thus allowing calculations when atomic resolution is not available or coarse-grained models are preferred. We parameterized the effective hydrodynamic radius of the elements in the atomic- and residue-level models using a very large set of experimental data for translational and rotational coefficients (intrinsic viscosity and radius of gyration) for >50 proteins. We also extended the calculations to very large proteins and macromolecular complexes, such as the whole 70S ribosome. We show that with proper parameterization, the two levels of resolution yield similar and rather good agreement with experimental data. The new version of HYDROPRO, in addition to considering various computational and modeling schemes, is far more efficient computationally and can be handled with the use of a graphical interface.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- García de la Torre J., Huertas M.L., Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. - PubMed
-
- Ortega A., García de la Torre J. Efficient, accurate calculation of rotational diffusion and NMR relaxation of globular proteins from atomic-level structures and approximate hydrodynamic calculations. J. Am. Chem. Soc. 2005;127:12764–12765. - PubMed
-
- García de la Torre J., del Río Echenique G., Ortega A. Improved calculation of rotational diffusion and intrinsic viscosity of bead models for macromolecules and nanoparticles. J. Phys. Chem. B. 2007;111:955–961. - PubMed
-
- Bloomfield V.A., Dalton W.O., Van Holde K.E. Frictional coefficients of multisubunit structures. I. Theory. Biopolymers. 1967;5:135–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

