Molecular call and response: the physiology of bacterial small RNAs
- PMID: 21843668
- PMCID: PMC3186873
- DOI: 10.1016/j.bbagrm.2011.07.013
Molecular call and response: the physiology of bacterial small RNAs
Abstract
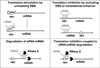
The vital role of bacterial small RNAs (sRNAs) in cellular regulation is now well-established. Although many diverse mechanisms by which sRNAs bring about changes in gene expression have been thoroughly described, comparatively less is known about their biological roles and effects on cell physiology. Nevertheless, for some sRNAs, insight has been gained into the intricate regulatory interplay that is required to sense external environmental and internal metabolic cues and turn them into physiological outcomes. Here, we review examples of regulation by selected sRNAs, emphasizing signals and regulators required for sRNA expression, sRNA regulatory targets, and the resulting consequences for the cell. We highlight sRNAs involved in regulation of the processes of iron homeostasis (RyhB, PrrF, and FsrA) and carbon metabolism (Spot 42, CyaR, and SgrS).
2011 Elsevier B.V. All rights reserved.
Figures

References
-
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biological Chemistry. 2011;392:277–289. - PubMed
-
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Current Opinion in Microbiology. 2007;10:156–163. - PubMed
-
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annual Review of Microbiology. 2010;64:475–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

