Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense-cored cell
- PMID: 21844238
- PMCID: PMC3257945
- DOI: 10.1128/IAI.05658-11
Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense-cored cell
Abstract
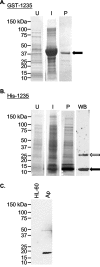
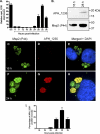
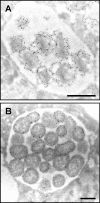
Anaplasma phagocytophilum is an obligate intracellular bacterium that invades neutrophils to cause the emerging infectious disease human granulocytic anaplasmosis. A. phagocytophilum undergoes a biphasic developmental cycle, transitioning between an infectious dense-cored cell (DC) and a noninfectious reticulate cell (RC). To gain insights into the organism's biology and pathogenesis during human myeloid cell infection, we conducted proteomic analyses on A. phagocytophilum organisms purified from HL-60 cells. A total of 324 proteins were unambiguously identified, thereby verifying 23.7% of the predicted A. phagocytophilum proteome. Fifty-three identified proteins had been previously annotated as hypothetical or conserved hypothetical. The second most abundant gene product, after the well-studied major surface protein 2 (P44), was the hitherto hypothetical protein APH_1235. APH_1235 homologs are found in other Anaplasma and Ehrlichia species but not in other bacteria. The aph_1235 RNA level is increased 70-fold in the DC form relative to that in the RC form. Transcriptional upregulation of and our ability to detect APH_1235 correlate with RC to DC transition, DC exit from host cells, and subsequent DC binding and entry during the next round of infection. Immunoelectron microscopy pronouncedly detects APH_1235 on DC organisms, while detection on RC bacteria minimally, at best, exceeds background. This work represents an extensive study of the A. phagocytophilum proteome, discerns the complement of proteins that is generated during survival within human myeloid cells, and identifies APH_1235 as the first known protein that is pronouncedly upregulated on the infectious DC form.
Figures


References
-
- Abdelrahman Y. M., Belland R. J. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959 - PubMed
-
- Borjesson D. L., et al. 2005. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J. Immunol. 174:6364–6372 - PubMed
-
- Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 - PubMed
-
- Carlyon J. A. 2005. Laboratory maintenance of Anaplasma phagocytophilum, p. 3A.2.1–3A.2.30 In Coico R., Kowalik T. F., Quarles J. M., Stevenson B., Taylor R.(ed.), Current protocols in microbiology. J. Wiley and Sons, Hoboken, NJ - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

