Architecture of human telomerase RNA
- PMID: 21844345
- PMCID: PMC3251123
- DOI: 10.1073/pnas.1100279108
Architecture of human telomerase RNA
Abstract
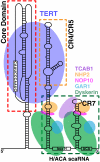
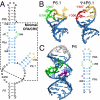
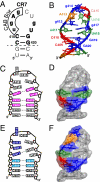
Telomerase is a unique reverse transcriptase that catalyzes the addition of telomere DNA repeats onto the 3' ends of linear chromosomes and plays a critical role in maintaining genome stability. Unlike other reverse transcriptases, telomerase is unique in that it is a ribonucleoprotein complex, where the RNA component [telomerase RNA (TR)] not only provides the template for the synthesis of telomere DNA repeats but also plays essential roles in catalysis, accumulation, TR 3'-end processing, localization, and holoenzyme assembly. Biochemical studies have identified TR elements essential for catalysis that share remarkably conserved secondary structures across different species as well as species-specific domains for other functions, paving the way for high-resolution structure determination of TRs. Over the past decade, structures of key elements from the core, conserved regions 4 and 5, and small Cajal body specific RNA domains of human TR have emerged, providing significant insights into the roles of these RNA elements in telomerase function. Structures of all helical elements of the core domain have been recently reported, providing the basis for a high-resolution model of the complete core domain. We review this progress to determine the overall architecture of human telomerase RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
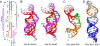
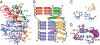

References
-
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. - PubMed
-
- Martínez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. - PubMed
-
- Wong JM, Collins K. Telomere maintenance and disease. Lancet. 2003;362:983–988. - PubMed
-
- Blasco MA. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. - PubMed
-
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

