Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism
- PMID: 21844365
- PMCID: PMC3182688
- DOI: 10.1073/pnas.1102164108
Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism
Abstract
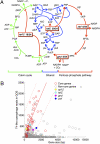
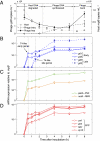
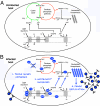
Cyanophages infecting the marine cyanobacteria Prochlorococcus and Synechococcus encode and express genes for the photosynthetic light reactions. Sequenced cyanophage genomes lack Calvin cycle genes, however, suggesting that photosynthetic energy harvested via phage proteins is not used for carbon fixation. We report here that cyanophages carry and express a Calvin cycle inhibitor, CP12, whose host homologue directs carbon flux from the Calvin cycle to the pentose phosphate pathway (PPP). Phage CP12 was coexpressed with phage genes involved in the light reactions, deoxynucleotide biosynthesis, and the PPP, including a transaldolase gene that is the most prevalent PPP gene in cyanophages. Phage transaldolase was purified to homogeneity from several strains and shown to be functional in vitro, suggesting that it might facilitate increased flux through this key reaction in the host PPP, augmenting production of NADPH and ribose 5-phosphate. Kinetic measurements of phage and host transaldolases revealed that the phage enzymes have k(cat)/K(m) values only approximately one third of the corresponding host enzymes. The lower efficiency of phage transaldolase may be a tradeoff for other selective advantages such as reduced gene size: we show that more than half of host-like cyanophage genes are significantly shorter than their host homologues. Consistent with decreased Calvin cycle activity and increased PPP and light reaction activity under infection, the host NADPH/NADP ratio increased two-fold in infected cells. We propose that phage-augmented NADPH production fuels deoxynucleotide biosynthesis for phage replication, and that the selection pressures molding phage genomes involve fitness advantages conferred through mobilization of host energy stores.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

