Dopamine D4 receptor, but not the ADHD-associated D4.7 variant, forms functional heteromers with the dopamine D2S receptor in the brain
- PMID: 21844870
- PMCID: PMC3219836
- DOI: 10.1038/mp.2011.93
Dopamine D4 receptor, but not the ADHD-associated D4.7 variant, forms functional heteromers with the dopamine D2S receptor in the brain
Abstract
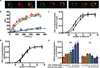
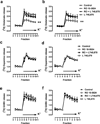
Polymorphic variants of the dopamine D(4) receptor have been consistently associated with attention-deficit hyperactivity disorder (ADHD). However, the functional significance of the risk polymorphism (variable number of tandem repeats in exon 3) is still unclear. Here, we show that whereas the most frequent 4-repeat (D(4.4)) and the 2-repeat (D(4.2)) variants form functional heteromers with the short isoform of the dopamine D(2) receptor (D(2S)), the 7-repeat risk allele (D(4.7)) does not. D(2) receptor activation in the D(2S)-D(4) receptor heteromer potentiates D(4) receptor-mediated MAPK signaling in transfected cells and in the striatum, which did not occur in cells expressing D(4.7) or in the striatum of knockin mutant mice carrying the 7 repeats of the human D(4.7) in the third intracellular loop of the D(4) receptor. In the striatum, D(4) receptors are localized in corticostriatal glutamatergic terminals, where they selectively modulate glutamatergic neurotransmission by interacting with D(2S) receptors. This interaction shows the same qualitative characteristics than the D(2S)-D(4) receptor heteromer-mediated mitogen-activated protein kinase (MAPK) signaling and D(2S) receptor activation potentiates D(4) receptor-mediated inhibition of striatal glutamate release. It is therefore postulated that dysfunctional D(2S)-D(4.7) heteromers may impair presynaptic dopaminergic control of corticostriatal glutamatergic neurotransmission and explain functional deficits associated with ADHD.
Conflict of interest statement
The authors declare no conflict of interest
Figures





References
-
- Lauzon NM, Laviolette SR. Dopamine D4-receptor modulation of cortical neuronal network activity and emotional processing: Implications for neuropsychiatric disorders. Behav Brain Res. 2010;208:12–22. - PubMed
-
- Tarazi FI, Campbell A, Yeghiayan SK, Baldessarini RJ. Localization of dopamine receptor subtypes in corpus striatum and nucleus accumbens septi of rat brain: comparison of D1-, D2-, and D4-like receptors. Neuroscience. 1998;83:169–176. - PubMed
-
- Svingos AL, Periasamy S, Pickel VM. Presynaptic dopamine D(4) receptor localization in the rat nucleus accumbens shell. Synapse. 2000;36:222–232. - PubMed
-
- LaHoste GJ, Swanson JM, Wigal SB, Glabe C, Wigal T, King N, et al. Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry. 1996;1:121–124. - PubMed
-
- Swanson JM, Kinsbourne M, Nigg J, Lanphear B, Stefanos GA, Volkow N, et al. Etiologic subtypes of attention-deficit/hyperactivity disorder: barin imaging, molecular genetics and environmental factors and the dopamine hypothesis. Neuropsychol Rev. 2007;17:39–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

