A data model and database for high-resolution pathology analytical image informatics
- PMID: 21845230
- PMCID: PMC3153692
- DOI: 10.4103/2153-3539.83192
A data model and database for high-resolution pathology analytical image informatics
Abstract
Background: The systematic analysis of imaged pathology specimens often results in a vast amount of morphological information at both the cellular and sub-cellular scales. While microscopy scanners and computerized analysis are capable of capturing and analyzing data rapidly, microscopy image data remain underutilized in research and clinical settings. One major obstacle which tends to reduce wider adoption of these new technologies throughout the clinical and scientific communities is the challenge of managing, querying, and integrating the vast amounts of data resulting from the analysis of large digital pathology datasets. This paper presents a data model, which addresses these challenges, and demonstrates its implementation in a relational database system.
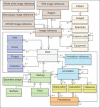
Context: This paper describes a data model, referred to as Pathology Analytic Imaging Standards (PAIS), and a database implementation, which are designed to support the data management and query requirements of detailed characterization of micro-anatomic morphology through many interrelated analysis pipelines on whole-slide images and tissue microarrays (TMAs).
Aims: (1) Development of a data model capable of efficiently representing and storing virtual slide related image, annotation, markup, and feature information. (2) Development of a database, based on the data model, capable of supporting queries for data retrieval based on analysis and image metadata, queries for comparison of results from different analyses, and spatial queries on segmented regions, features, and classified objects.
Settings and design: The work described in this paper is motivated by the challenges associated with characterization of micro-scale features for comparative and correlative analyses involving whole-slides tissue images and TMAs. Technologies for digitizing tissues have advanced significantly in the past decade. Slide scanners are capable of producing high-magnification, high-resolution images from whole slides and TMAs within several minutes. Hence, it is becoming increasingly feasible for basic, clinical, and translational research studies to produce thousands of whole-slide images. Systematic analysis of these large datasets requires efficient data management support for representing and indexing results from hundreds of interrelated analyses generating very large volumes of quantifications such as shape and texture and of classifications of the quantified features.
Materials and methods: We have designed a data model and a database to address the data management requirements of detailed characterization of micro-anatomic morphology through many interrelated analysis pipelines. The data model represents virtual slide related image, annotation, markup and feature information. The database supports a wide range of metadata and spatial queries on images, annotations, markups, and features.
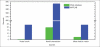
Results: We currently have three databases running on a Dell PowerEdge T410 server with CentOS 5.5 Linux operating system. The database server is IBM DB2 Enterprise Edition 9.7.2. The set of databases consists of 1) a TMA database containing image analysis results from 4740 cases of breast cancer, with 641 MB storage size; 2) an algorithm validation database, which stores markups and annotations from two segmentation algorithms and two parameter sets on 18 selected slides, with 66 GB storage size; and 3) an in silico brain tumor study database comprising results from 307 TCGA slides, with 365 GB storage size. The latter two databases also contain human-generated annotations and markups for regions and nuclei.
Conclusions: Modeling and managing pathology image analysis results in a database provide immediate benefits on the value and usability of data in a research study. The database provides powerful query capabilities, which are otherwise difficult or cumbersome to support by other approaches such as programming languages. Standardized, semantic annotated data representation and interfaces also make it possible to more efficiently share image data and analysis results.
Keywords: Data models; databases; digitized slides; image analysis.
Figures

Similar articles
-
Managing and Querying Whole Slide Images.Proc SPIE Int Soc Opt Eng. 2012 Feb 16;8319:83190J. doi: 10.1117/12.912388. Proc SPIE Int Soc Opt Eng. 2012. PMID: 22844574 Free PMC article.
-
A high-performance spatial database based approach for pathology imaging algorithm evaluation.J Pathol Inform. 2013 Mar 14;4:5. doi: 10.4103/2153-3539.108543. Print 2013. J Pathol Inform. 2013. PMID: 23599905 Free PMC article.
-
Towards Building a High Performance Spatial Query System for Large Scale Medical Imaging Data.Proc ACM SIGSPATIAL Int Conf Adv Inf. 2012 Nov 6;2012:309-318. doi: 10.1145/2424321.2424361. Proc ACM SIGSPATIAL Int Conf Adv Inf. 2012. PMID: 24501719 Free PMC article.
-
Applications and challenges of digital pathology and whole slide imaging.Biotech Histochem. 2015 Jul;90(5):341-7. doi: 10.3109/10520295.2015.1044566. Epub 2015 May 15. Biotech Histochem. 2015. PMID: 25978139 Review.
-
Image analysis and machine learning in digital pathology: Challenges and opportunities.Med Image Anal. 2016 Oct;33:170-175. doi: 10.1016/j.media.2016.06.037. Epub 2016 Jul 4. Med Image Anal. 2016. PMID: 27423409 Free PMC article. Review.
Cited by
-
Accelerating Pathology Image Data Cross-Comparison on CPU-GPU Hybrid Systems.Proceedings VLDB Endowment. 2012 Jul;5(11):1543-1554. doi: 10.14778/2350229.2350268. Proceedings VLDB Endowment. 2012. PMID: 23355955 Free PMC article.
-
Multiscale integration of -omic, imaging, and clinical data in biomedical informatics.IEEE Rev Biomed Eng. 2012;5:74-87. doi: 10.1109/RBME.2012.2212427. IEEE Rev Biomed Eng. 2012. PMID: 23231990 Free PMC article. Review.
-
Big Data: the challenge for small research groups in the era of cancer genomics.Br J Cancer. 2015 Nov 17;113(10):1405-12. doi: 10.1038/bjc.2015.341. Epub 2015 Oct 22. Br J Cancer. 2015. PMID: 26492224 Free PMC article. Review.
-
Scalable analysis of Big pathology image data cohorts using efficient methods and high-performance computing strategies.BMC Bioinformatics. 2015 Dec 1;16:399. doi: 10.1186/s12859-015-0831-6. BMC Bioinformatics. 2015. PMID: 26627175 Free PMC article.
-
Managing and Querying Whole Slide Images.Proc SPIE Int Soc Opt Eng. 2012 Feb 16;8319:83190J. doi: 10.1117/12.912388. Proc SPIE Int Soc Opt Eng. 2012. PMID: 22844574 Free PMC article.
References
-
- Furness PN, Taub N, Assmann KJ, Banfi G, Cosyns JP, Dorman AM, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol. 2003;27:805–10. - PubMed
-
- Saltz J, Kurc T, Cholleti S, Kong J, Moreno C, Sharma A, et al. Proceedings of the Annual Symposium of American Medical Informatics Association Summit on Translational Bioinformatics (AMIA-TBI 2010) San Francisco, LA: 2010. Mar, Multi-scale, integrative study of brain tumor: In silico brain tumor research center.
-
- Catalyurek UV, Beynon MD, Chang C, Kurc TM, Sussman A, Saltz JH. The virtual microscope. IEEE Trans Inf Technol Biomed. 2003;7:230–48. - PubMed
Grants and funding
- RC4 MD005964/MD/NIMHD NIH HHS/United States
- P30 CA072720/CA/NCI NIH HHS/United States
- R01 LM009239/LM/NLM NIH HHS/United States
- R01 CA156386/CA/NCI NIH HHS/United States
- P20 EB000591/EB/NIBIB NIH HHS/United States
- N01 CO012400/CA/NCI NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- U54 CA113001/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R01 LM011119/LM/NLM NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- UL1 RR025008/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

