Quantitative characterization of temperature-independent and temperature-dependent protein-protein interactions in highly nonideal solutions
- PMID: 21846103
- PMCID: PMC3488770
- DOI: 10.1021/jp2049266
Quantitative characterization of temperature-independent and temperature-dependent protein-protein interactions in highly nonideal solutions
Abstract
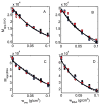
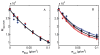
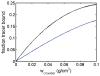
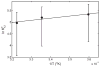
The interaction among each of three dilute "tracer" proteins (bovine serum albumin, superoxide dismutase, and ovomucoid) at a concentration of 2 mg/mL and each of two "crowder" proteins (ovomucoid and BSA) at concentrations up to 100 mg/mL was characterized by analysis of dependence of the equilibrium gradients of both tracer and crowder upon the concentration of crowder. The equilibrium gradients of both crowder proteins were found to be independent of temperature over the range 5-37 °C. The equilibrium gradients of tracer BSA and ovomucoid in the complementary crowder species were likewise found to be independent of temperature over this range, indicating that interaction among these tracers and crowders is predominantly repulsive and essentially entirely entropic in nature. The equilibrium gradient of tracer SOD in BSA was also found to be independent of temperature over this range, but the gradient of tracer SOD in ovomucoid depended significantly upon temperature in a manner indicating a significant enthalpic (attractive) component of the overall interaction between SOD and ovomucoid. The experimental data are analyzed using model-free expansions of the thermodynamic activity coefficients of tracer and crowder in powers of the concentration of crowder and using approximate statistical thermodynamic models based upon highly simplified descriptions of molecular structure and interactions. Detailed analysis of the results indicates a relatively small contribution of nonspecific attraction to the total protein-protein interaction, which is dominated by steric repulsion.
© 2011 American Chemical Society
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

